The Prebys Center conducts drug discovery research in diverse disease areas such as autoimmunity, cardiovascular and metabolic diseases, cancer and neurodegenerative disorders.
Our drug discovery process involves assembling teams with cross disciplinary expertise, who leverage the diverse capabilities of the center, deploying project management to organize activities around a milestone driven process aimed at ultimately discovering a new drug. A brief description of these capabilities is described below, and followed by some current and past examples of our research productivity.
The Prebys Center represents an investment of more than $20M in capital equipment, over 12,500 sq. ft. of dedicated space, and supplies key supporting functions for drug discovery including:
- Highly experienced assay development & HTS teams
- State-of-art robotic HTS & high-content screening operations
- Diverse chemical libraries (>900,000 cpds), >85% of FDA drugs
- Diverse screening platforms – phenotypic HCS / biophysical
- Integrated drug discovery teams (biologists & medicinal chemists)
- In vitro in vivo pharmacology (ADME/PK)
- Cell biology team focused on stem cell derived models of disease
- Experienced outsourcing office (chemistry/PK/Tox)
- Extensive hit-to-lead & lead optimization experience & expertise
- Portfolio & project management
Drug Discovery Technology
Assay Development
| Prebys Center HTS assay platforms 384/1536 well format | Biochemical and Biophysical Assays | Cell Based Assays | ||||||
|---|---|---|---|---|---|---|---|---|
| Enzyme activity | Protein Thermal Shift | Complex displacement | Non-phenotypic | Phenotypic | ||||
| Binding (PCA) | Reporter | Cellular Thermal Shift | qPCR | Image or High Content | ||||
Over the past 15 years the Prebys Center has established a broad array of diverse assay platforms suitable for compound screening in a 384/1536 well format. For biochemical and biophysical assays these include classic enzyme activity assays, protein thermal shift assays and an array of complex displacement assays. For cell-based assays these include commonly used binding assays (PCA), a diversity of reporter assays, Cellular thermal shift (e.g. split luciferase), 4 color qPCR assays as well as phenotypic microscopy-based assays, often referred to as high content assay, described in the section on high content imaging platforms. We use biologically relevant and agnostic approaches, developing the best assay for each specific target. In addition to know-how, the team is supported by cutting edge technology, such as label-free EPIC, protein thermal shift assays in multiple formats and split luciferase assays that enable in cell target engagement by compounds to be monitored.
Cell Biology
The Cell Biology group is focused on development of patient cell specific and human induced pluripotent stem cell (hiPSC)-based disease models for drug screening and target identification. They aim to design human cell-based assays that reflect higher order cellular functions and recapitulate disease phenotypes, yet have the throughput and reproducibility required for drug discovery. The group has a focus on establishing model systems of brain circuitry with hiPSC derived cells using phenotypic microscopy-based assays as well as 48/96 well multiple electrode array (MEA) instrumentation to monitor neuronal firing.
Cheminformatics
The Cheminformatics group applies cheminformatics tools, computational chemistry, and computer-aided molecular design approaches to support various aspects of preclinical drug discovery research. In support of assay development, HTS, and HCS activities, the Cheminformatics group provides databases and tools for compound registration, inventory management, plate formatting and tracking, data entry and organization, data mining, and reporting. The database infrastructure is supported with a user-friendly interface and provides access to the data from every assay run at the institute. The compound libraries supported with PAINs filters, sophisticated structure-alerts, and compound similarity algorithms for library design, library diversity analysis, and rapid identification of the most promising hits. HTS bioactivity is tracked and annotated to identify promiscuous compounds and known mechanisms of action. Drugs and known bioactive molecules from our specialty libraries are also annotated for mechanism and therapeutic effect.
High Content Imaging Platforms
The High Content Imaging (HCI) Platforms group performs assay development, screening, and data analysis/mining for high content screens, where the readout is based on images obtained using high-throughput microscopy systems. In fact, the Prebys Center has developed over 50 HCS assays, many using immunofluorescence markers against endogenous proteins, in 384- and 1,536-well formats. More than 20 of these assays have been successfully converted to large-scale HC screens and demonstrate robustness typically required for both Hit Identification as well as SAR support. The HCI Platforms scientists provide expertise in all areas of high content image-based screens including microscopy instrumentation, assay biology and assay design/proof-of-concept, sample preparation, image acquisition, image analysis, image data management, data mining, and algorithm development. Developed assays range from detection of β-arrestin-GFP GPCR activity, to cell shape and cytoskeletal rearrangement and complex multiplexed, multi-parametric kidney podocyte or cell health profiling assays, and include advanced methods such as FRET imaging, 3D spheroid analyses, and cell migration assays.
The HCI Platforms team is also responsible for implementation, maintenance, and enhancements of the complex HCI platform infrastructure and workflows, which enable routine screening of small (<1,000) to large-scale (>100,000) chemical compound libraries using image-based phenotypic assays. The HCI infrastructure includes several confocal and/or widefield imaging systems from 4x to 60x magnifications with high resolution objectives of up to 1.2 numerical aperture. Our HCI team has created numerous complex assay algorithms for rapid batch processing during large-scale HCS while also providing easy-to-use access to standard image analysis tools and workflows. They have also designed and implemented integrated automated microscopy systems with advanced photonics and robotics capabilities for automated use in both fixed and live-cell formats, including a robotic system enabling high-throughput multi-day time-lapse imaging experiments.
High Throughput Screening and Compound Management
HTS capabilities are fundamental to the small molecule drug discovery efforts of the Prebys Center, with over 300 HTS screening campaigns successfully completed to date. The facilities and expertise of the HTS team enabled it to partner with investigators to conduct large chemical library screens, typically >300,000 compounds, with the goal of identifying hits or leads that represent the starting points for small molecule drug discovery programs. The group has developed highly advanced automation infrastructure including state-of-the-art liquid handling devices, multiple robotic integration systems as well as a diverse range of signal detection instruments.
A major portion of the HTS screening lab is devoted to housing three fully-integrated robotic HTS systems, liquid handler workstations, various plate readers, and one integrated image-based high-content screening system. The centerpiece of the screening facility is a HighRes Biosolutions system POD-1, this has a central robotic arm surrounded by docking stations that accept universal cart on to instrumentation is placed. This system offers unprecedented modularity, greatly extending the flexibility, utility, and lifespan of the system. Echo 555 instruments are used as the workhorse liquid handling units, able to perform contact-free transfer of 2.5 nL droplets directly from a source to a destination plate using focused energy of acoustic waveforms. As a fully integrated component of the HiRes system, acoustic transfer of compounds from the on-line compound plates to the assay plates for primary screening, as well as for single-concentration and dose-response hit picking and confirmation, can be achieved with an overall maximum throughput of >300,000 wells/day. The activities of the HTS team are complimented and aided by the Compound Management team who deploy several automated sample management and liquid-handling instruments. Taken together this enables scientists to screen an array of diverse and large compound libraries.

Sanford Burnham Prebys
Diversity Libraries
Compound Management
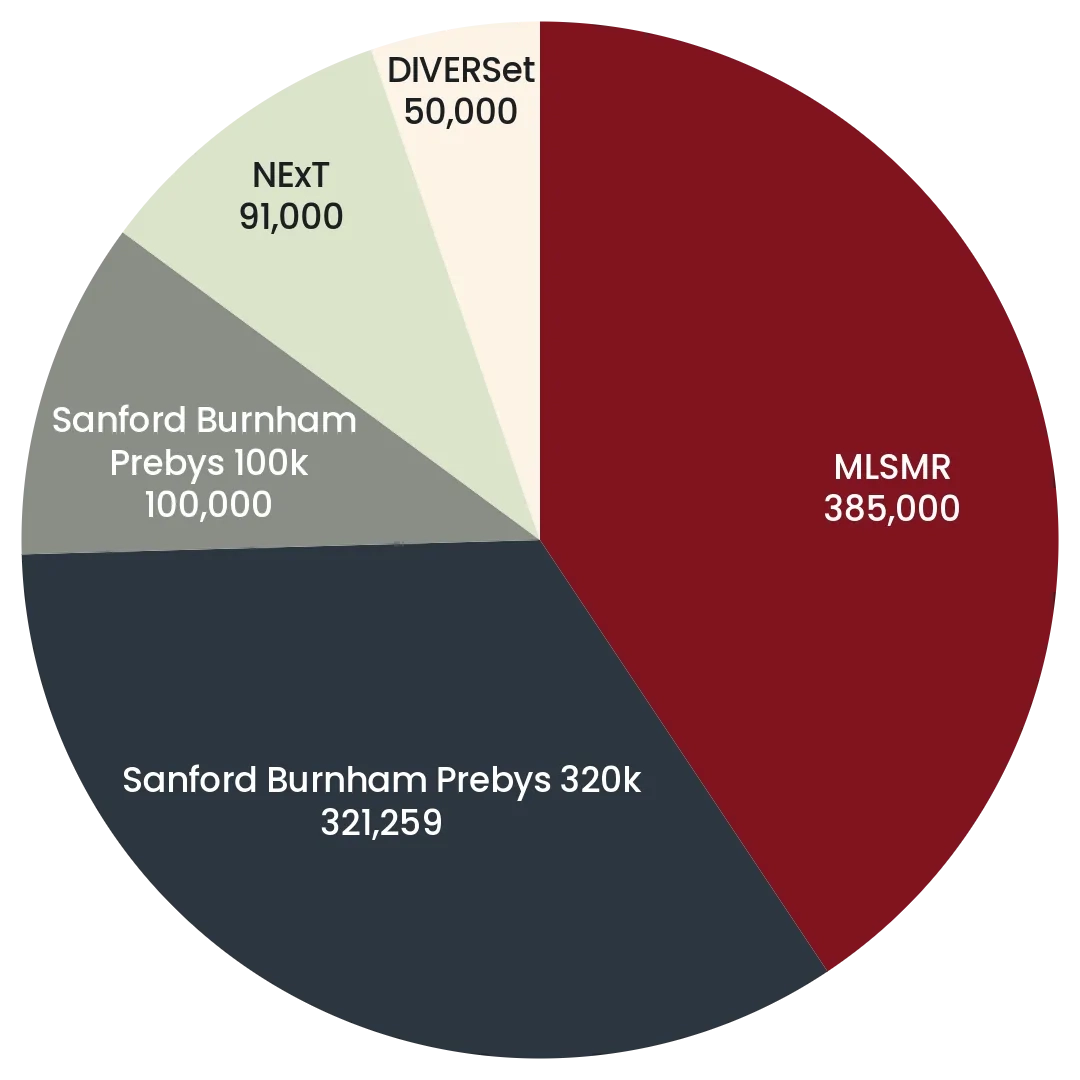
The Prebys Center has multiple large diverse chemical libraries that collectively comprise >1,000,000 compounds in a format suitable for high throughput robotic screening. The pie charts below summarize the relative sizes of the main components the screening collections comprising, in addition the center has assembled smaller focused library collections comprising >50,000 compounds, including a non redundant set of 4,200 drugs.
In vitro Pharmacology (Bioanalytical/DMPK)
The in vitro Pharmacology group excels at quickly providing high quality results to the medicinal chemistry group in their drug development efforts. Having direct and immediate access to our scientific expertise is what sets us apart from contract research organizations and other service providers. The bioanalytical facility has the capability to perform a number of critical drug discovery ADME (Absorption, Distribution, Metabolism, Excretion) assays including microsomal and plasma stability, plasma protein and tissue binding experiments, solubility, PAMPA (Parallel Artificial Membrane Permeability Assay), Caco2 permeability, Cytochrome P450 inhibition and metabolite profiling. It also has the capacity to analyze samples from in-house or external in vivo PK and PD experiments. Sample processing is enhanced by the use of a Biotek Precision 2000 liquid handler for reliably preparing standard calibration and QC standard dilution curves as well as an Apricot Designs PP-550 96 well plate liquid handler into the work flow. Analysis of drug levels and metabolite profiling is by LC-MS/MS using a Shimadzu 20AD HPLC with a Nexera SIL-30ACmp six plate autosampler coupled to an AB SCIEX API 4000 QTRAP® LC/MS/MS Systems.
| Preclinical Capabilities: AMDE/PK – assays available in two tiers | |||
|---|---|---|---|
| Tier 1 in vitro (least costly) | Aqueous Solubility | In vitro Safety and Toxicology (more costly) | Ames Test |
| Microsomal Stability | CEREP Profiling | ||
| PAMPA (Permeability) | Micronuclease Studies | ||
| PAMPA-BBB (Permeability) | Mouse Lymphoma Assay | ||
| Plasma Stability | hERG Inhibition | ||
| Plasma Protein Binding | in vitro cardio-tox | ||
| Tier 2 in vitro (costly) | Cytochrome P450 Inhibition | In vivo Analysis (more costly) | Preliminary Formulation |
| Cytochrome P450 Induction | CNS Penetration | ||
| Cellular Toxicity | Tissue Distribution | ||
| Caco-2 Permeability | Studies | ||
| MDR1-MDCK Permeability | Bioanalytical Method Development | ||
| Pgp transporters | IV/IP/PO Pharmacokinetics | ||
| Metabolite Profiling | RACE (Rapid Assessment of Compound Exposure) | ||
Medicinal Chemistry
The Medicinal Chemistry team has extensive knowledge and experience in all aspects of drug discovery, including hit selection, initial SAR exploration and lead optimization. They initiate the discovery of lead structures from hits identified high-throughput screening (Hit-to-Lead chemistry), known literature compounds, and de novo design. Subsequent structure-activity relationship (SAR) studies around the lead compounds are performed to maximize potency and efficacy, while simultaneously optimizing for selectivity, pharmacokinetics, toxicity, and drug like properties such as solubility and permeability. Over the past 5 years the team has designed, synthesized and characterized about 2,000 novel compounds each year. With the support of the Institute, a significant effort has been made to acquire the latest in cutting edge synthesis technologies, enabling very rapid production of high-quality small molecule libraries. The lab houses a CEM automated microwave synthesizer for additional synthetic capabilities. Multiple alternatives exist for the automated purification of final library compounds. Overall the team has full analytical capabilities (LC-MS and NMR), and can re-purify, re-synthesize and scale up (to 50-100g) of lead compounds. The team spearheads candidate compound selection and optimization and is integrated with the DMPK group, which performs in vitro ADME assays, including plasma and microsomal stability, protein binding, solubility and permeability, as well as in vivo PK.
Exploratory Pharmacology (Bioanalytical/DMPK)
The Exploratory Pharmacology group provides rapid, high-quality assessments of drug-like properties to support informed decision-making in drug development. Leveraging scientific expertise, optimized workflow and state-of-the-art infrastructure, we enhance efficiency across the drug discovery pipeline.
| Tier 1 ADME In vitro | Tier 2 ADME In vitro | Drug-like Properties In vivo |
|---|---|---|
| Kinetic Solubility | Cyp P450 inhibition | Analytical method development |
| Thermodynamic solubility | In vitro hERG binding | IV/IP/PO Pharmacokinetics |
| Microsomal Stability | Metabolite profiling | Oral bioavailability |
| Hepatocyte Stability | Plasma Protein Binding | Formulation screening |
| Plasma Stability | Tissue free fraction | Tissue/CNS Distribution |
Comprehensive ADME Profiling
- ADME Assays: Solubility, microsomal and hepatocyte (mouse, rat, monkey, human, minipig, dog and others on demand) stability, plasma protein and tissue binding, permeability, Cytochrome P450 inhibition, hERG binding, and metabolite profiling.
- Bioanalytical Capabilities: Utilizing state-of-the-art Triple Quadrupole mass spectrometry (API 6500+ QTRAP) for ultra-sensitive drug quantification in small sample volumes with limit of quantifications typically below 1nM.
Efficient Micro-PK Study
- Micro-sampling Techniques: Small sample size requirements for sensitive analytical equipment in combination with Saphenous, retro-orbital, and terminal bleeding methods allow for serial blood sampling from mice, generating complete PK profiles with minimal animal use.
- Routes of Administration: IV, IP, PO, SC, and topical routes are conducted in mice and rats.
Extended Services
- External Study Support: Analysis of plasma/tissue samples from external PK/PD studies using LC-MS/MS.
- Pharmacological Evaluations: Formulation screening, allometric scaling, and AI/ML-driven predictions for pharmacological properties (for academic clients/collaborators).
- We also have access to reliable vendors who perform high quality studies including permeability, CYP induction, met-ID, genotoxicity and other assays.
By integrating cutting-edge technology with specialized expertise, we accelerate drug candidate identification while adhering to ethical and cost-effective practices.
Protein Production and Analysis
Custom Recombinant Protein Production
The Protein Production and Analysis group generates custom protein constructs and expresses and purifies recombinant proteins for variety of drug discovery projects including cell-based and protein-based chemical library screening, assay development, hit conformation, characterization of drug candidate – target interaction, etc. Production is carried out in bacterial (E. coli), insect/baculovirus and mammalian expression systems, and involves cloning of target genes; generation cell lines/viruses; optimization of protein expression; development of protein purification protocols; and protein purification from large scale cell culture.
Biophysical Protein Analysis
Protein targets are characterized by variety of biophysical methods including analytical ultracentrifugation (AUC), differential scanning calorimetry (DSC), isothermal titration calorimetry(ITC), microscale thermophoresis (MST) and fast kinetics/stopped-flow. Data obtained from biophysical analysis are used for quality control of protein preparations, to assist design and troubleshooting of HTS assays, to confirm binding of HTS hits, and to guide structural biology(crystallization, NMR) drug discovery effort.
Project Management Capabilities
The Prebys Center has a team of dedicated, trained project managers who are experienced in small molecule drug discovery. We work with our internal and external project team members and collaborators to ensure clarity on objectives, critical path activities and interdependencies of a project plan. The team also provides resource management to ensure delivery of projects on time and on budget. We understand the importance of project governance and facilitate these processes both internally and externally with our partners to ensure open communication and expectations in decision making.
Structure-Guided Drug Design
Crystallography
The Prebys Center has Beckman Biomek FX, Agilent Bravo, and TTP Labtech Mosquito liquid handling platforms to facilitate automated execution of protein crystallization trials. It also houses a Formulatrix Rock Imager 2 for evaluation of such trials. SBP’s Crystallography Core has additional supporting equipment including a Phoenix Microdrop Liquid Handling system and two Formulatrix incubators (4°C and 20°C) with automated image analysis to detect crystal growth. For diffraction analysis, the Core has a Rigaku FR-E SuperBright Ultra high-intensity rotating anode generator supplying two independent diffraction stations equipped with X-stream cryogenic systems. These two stations’ detectors are an R-Axis HTC three-image plate detector and an R-Axis IV two-image plate detector. The Center also has access to SSRL’s macromolecular crystallography beamlines to enable acquisition of very high-quality data and prosecution of challenging targets which produce poorly diffracting crystals.
Computational Chemistry
The SGDD group employs MOE (Molecular Operating Environment), an integrated computer-aided molecular design platform to analyze structural data, create homology models, run molecular dynamics simulations, and calculate conformational ensembles. We are fully enabled to perform drug discovery campaigns using both structure-based design and ligand-based design, which accelerate series development, by providing guidance for ligand optimization and de novo ligand discovery. The suite also provides a platform for in silico Virtual Screening. We employ both MOE and the ChemAxon collections of physical-chemical predictors and calculators to drive the optimization of drug-like properties and develop quantitative structure-activity relationships and structure property relationships (QSAR/QSPR)
Active Drug Discovery / Translational Sciences Grants / Contracts
Neurological Disorders and Neurodegeneration Grant Awards
- Development of SBI-553, an allosteric modulator of NTR1, for the treatment of substance use disorders
- Discovery of selective allosteric inhibitors for the EphA4 kinase (Alzheimer’s disease)
- Novel antagonists to the N-terminal Domain of CRF Receptor Type 1 for Alzheimer’s Disease
- Managing L-DOPA Induced Dyskinesias in Parkinson’s disease by Targeting Dopamine D1 Receptor Biased Signaling
- Targeting Neuroinflammation in Alzheimer’s with Novel CX3CR1 Agonists
- Development of Ciliary Neurotrophic Factor Receptor Agonists for ALS Therapy
- High Throughput screen to discover modulators of TREM2 activity (Alzheimer’s disease)
- High-throughput Screen for Discovery of Ciliary Neurotrophic Factor Receptor Agonists for the Treatment of Motor Neuron Diseases
- iPSC-based platform development for major psychiatric disorder modeling and discovery National Cooperative Reprogrammed Cell Research Groups to Study Mental Illness
- Development of STEP allosteric Inhibitors as Novel Therapeutics for Alzheimer’s Disease
- Farnesyl Transferase Inhibitors to Treat Tauopathies
Oncology Grant Awards
- Optimization and Characterization of “MYC Degraders” for Pediatric Medulloblastoma
- Development of Novel Cyclophilin B/PIKfyve Inhibitors for Treatment of Malignant Peripheral Nerve Sheath Tumors
- Defining TAO3 as a therapeutic target in melanoma
- Identification of Small Molecule Modulators of Mitochondrial Creatine Kinase 1
- GOLPH3 pathway inhibitors for cancer.
- Allosteric inhibitors of SHP2 for treatment of cancer.
- A screen for small Molecule Inhibitors of CCF to Suppress Senescence-Associated Inflammation
- Allosteric Inhibitors of PHGDH, A Cancer Metabolism Target.
- Early development of small molecule Dendritic cell Immunopotentiators for the treatment of solid tumors
- High Throughput Screening to Discover Chemical Inhibitors of Quiescin Sulfhydryl Oxidase 1
Infectious and Inflammatory Diseases Grant Awards
- Development and Advancement of Broad-spectrum Respiratory Antivirals
- Large-scale screen with a novel assay against RNA editing to identify anti-trypanosomal agents
- Brain pathology and function in a chronic mouse model of ZIKV transmission
- HTS Targeting HIV-1 Protease auto-processing for First in Class Drug Discovery
- Aging, Polypharmacy, and Neurotoxicity in Adults Living with HIV
Cardiovascular and Metabolic Diseases Grant Awards
- The development of TGR5 antagonists for the treatment of cholangiopathies
- Small molecule inhibitors of LMPTP: an obesity drug target
- Activation of the Secretin receptor as a strategy for the treatment of heart failure
- Universal positive allosteric modulators of CCK1R without intrinsic agonist activity for the treatment of obesity
- Development of allosteric HIPK4 inhibitors as non-hormonal male contraceptives
- Reversing Pulmonary Fibrosis Through Dopamine D1 Receptor Agonism
- Podocyte-based HCS assays for discovering therapeutics against kidney diseases
Our Research
The Prebys Center is a fully capable early drug discovery research operation with a strong track record of collaboration, high grant success rates and productive research. The following programs have advanced chemical leads:
Neuroscience
- NTR1
- NAMPT
- GR receptor passive antagonist
Oncology
- ULK1
- ENPP1
- Myc degrader
- eIF4g
Metabolism and Cardiovascular Disease
- TNAP
- APJ
- LMPTP
- TGR5
Immunology and Infectious Disease
- EBI2
- Plasmodium f. G6PD
