In a recent collaborative research study between two brothers—one a rheumatologist and the other a medical engineer—novel shaped nanoparticles were able to deliver anti-osteoarthritis drugs directly to the cells that drive the onset and progression of osteoarthritis (OA). The findings show promise to improve the treatment options for the nearly 21 million Americans, 25 years of age and older, that suffer from this chronic, often debilitating disease.
“We are excited to have developed nanoparticles which can efficiently and safely bring anti-OA drugs into the cells called chondrocytes that cause OA,” said Massimo Bottini, Ph.D., adjunct assistant professor in the Bioinformatics and Structural Biology Program at Sanford-Burnham. “Our method not only delivered the drug effectively, but stayed in the joint for a prolonged time without causing side effects. This is a significant improvement over previous attempts to deliver anti-OA drugs to affected joints.”
About osteoarthritis Under normal conditions, the extracellular matrix of the joint is maintained through a continual remodeling process in which low levels of different enzymes that produce and degrade cartilage are maintained. However, with increasing age and general wear and tear on joints, OA can occur when the enzymes that degrade cartilage are overproduced, creating an imbalance that leans toward the loss of collagen and joint impairment.
“For advanced OA, joint-replacement surgery is the only option for patients to regain comfortable and normal joint functions. For less severe cases, there is currently no medical therapy that can slow down or halt progression of the disease. This makes OA one of the largest unmet clinical needs in the field of rheumatology,” said Nunzio Bottini, MD, Ph.D., an associate professor in the Division of Cellular Biology at the La Jolla Institute for Allergy and Immunology (LIAI), who is also a practicing rheumatologist—and Massimo’s brother.
“The goal of treating OA is to restore the balance of the enzymes that control the matrix environment. Since there is no blood supply to the joint, drugs to treat the disease must be injected directly into the joint,” said Nunzio.
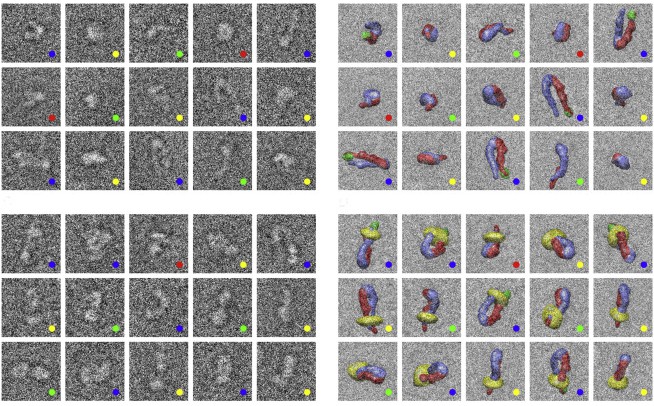
“Until now, scientists have tried using spherical nanoparticles to deliver anti-OA drugs. But the physical shape and size of the spheres predisposes them to diffuse into the synovial fluid and be flushed out of the joint before they can be effective,” said Massimo. “We have designed a one-dimensional linear nanoparticle made of graphite that is 100,000 times thinner than a human hair. This unique nanoparticle is engineered to travel through the negatively charged extracellular matrix and carry molecules to the nucleus of chondrocytes to turn off the genes that cause the disease.”
The study Using a mouse model for OA, the brothers injected the novel nanoparticle loaded with a gene inhibitor into the knees of affected mice. The nanoparticle delivered large amounts of the gene inhibitor to the cytoplasm and the nucleus of chondrocytes. Importantly, particles remained in the joint for two weeks compared to only few days for spherical nanoparticles.
“This is a significant improvement over previous attempts to deliver drugs to OA joints,” said Massimo. “Our next step is to further optimize the nanoparticle, see how long it remains in the body, and move to clinical studies in humans,” said Massimo.
“Arthritis is a complex disease and integrated work between technologists—such as my brother Massimo—and biologists like me significantly increases the chance to make major treatment advances. Our next objective is to secure NIH funding to continue applying our complementary expertise to the quest to improve the lives of those suffering from arthritis,” added Nunzio.
The collaborative study was published in ACS Nano and performed at both Sanford-Burnham and LIAI. Cristiano Sacchetti, PhD, a shared postdoctoral fellow in Massimo and Nunzio Bottini’s laboratories was lead author on the paper.
A link to the paper can be found at: http://pubs.acs.org/doi/abs/10.1021/nn504537b.