Michael Karin received a PhD in Molecular Biology from UCLA in 1979 and conducted postdoctoral research with Prof. Beatrice Mintz at Fox Chase Cancer Center from 1979 to 1980 and with Prof. John Baxter at UCSF from 1980 to 1983. He served as an Assistant Professor in the Department of Microbiology at USC School of Medicine from 1983 to 1985 and, in 1986, joined the Department of Pharmacology at UCSD School of Medicine, where he rose through the ranks to become a Distinguished Professor until his retirement in 2024. That same year, Karin joined the Sanford Burnham Prebys Medical Research Institute as the Director of the Center for Metabolic and Liver Diseases.
Throughout his academic career, Karin investigated stress and inflammation signaling, employing a full range of experimental approaches from biochemistry and molecular cell biology to animal pathophysiology. By discovering how infection, inflammation, radiation, or environmental stressors activate AP-1, NF-κB, and other transcription factors, he explored how these processes contribute to the pathogenesis of cancer, as well as degenerative and metabolic diseases. His group elucidated fundamental mechanisms by which inflammation and obesity promote tumor development and contribute to type II diabetes and insulin resistance, and was among the first to highlight the role of inflammation in metabolic disease.
Karin’s group established how members of the IL-6 cytokine family control the development of colorectal (CRC) and liver (HCC) cancers through activation of STAT3 and YAP, and identified cell type–specific mechanisms by which NF-κB activation via IκB kinases (IKK)—which his group was the first to discover—affects the pathogenesis of various cancers. His research also demonstrated that not only innate immune cells, such as macrophages, but also adaptive immune cells, including T regulatory cells and B lymphocytes, contribute to tumorigenesis. This body of work helped to establish the field of Inflammation and Cancer.
More recently, Karin’s group demonstrated the existence of immunosuppressive plasma cells and their role in negatively regulating immunosurveillance and immunotherapy. His current work focuses on HCC, CRC, and pancreatic ductal adenocarcinoma (PDAC). The team has developed robust mouse models for studying non-alcoholic steatohepatitis (NASH)-induced HCC and chronic pancreatitis–accelerated PDAC, using them to discover how adaptive immunity controls HCC development and to investigate the role of the autophagy substrate p62 in inflammation and HCC/PDAC pathogenesis.
They discovered that activation of the p62–KEAP1–NRF2 cascade confers resistance to autophagy inhibitors, leading to the development of a new PDAC and HCC therapeutic approach targeting autophagy and macropinocytosis. These mouse models have also been used to study how metabolism and nutrition affect NASH and HCC development, with a focus on the adverse effects of fructose and alcohol intake and exposure to toxicants.
Board Appointments, Conference Organization or Other Memberships
National Academy of Sciences, 2005
National Academy of Medicine, 2011
Associate member of the European Molecular Biology Association, 2007
Honors and Recognition
American Association for Cancer Research (AACR) G.H.A. Clowes Award for Outstanding Basic Cancer Research, 2020
William B. Coley Award for Distinguished Research in Basic and Tumor Immunology, 2013
Charles Rodolphe Brupbacher Prize for Cancer, 2013
Harvey Prize for Human Health, 2010
Morton I. Grossman Lectureship, 2003
Education
PhD, University of California Los Angeles, Molecular Biology, 1979
Postdoctoral fellowship, Fox Chase Cancer Center, 1980
Postdoctoral fellowship, University of California San Francisco, 1983
Related Disease
Cancer, Liver Cancer, Liver Diseases, Metabolic Diseases
Research in the Karin lab is currently focused on three general research areas: 1. Inflammation biology and how it is modulated by the individual metabolic and nutritional status. 2. Inflammation and metabolic disease, such as type 2 diabetes and metabolic dysfunction associated steatotic liver disease (MASLD). 3. The role of inflammation in cancer, including pancreatic, liver and gastrointestinal cancers. We have also started investigating the role of interorgan communication in metabolic disease and cancer.
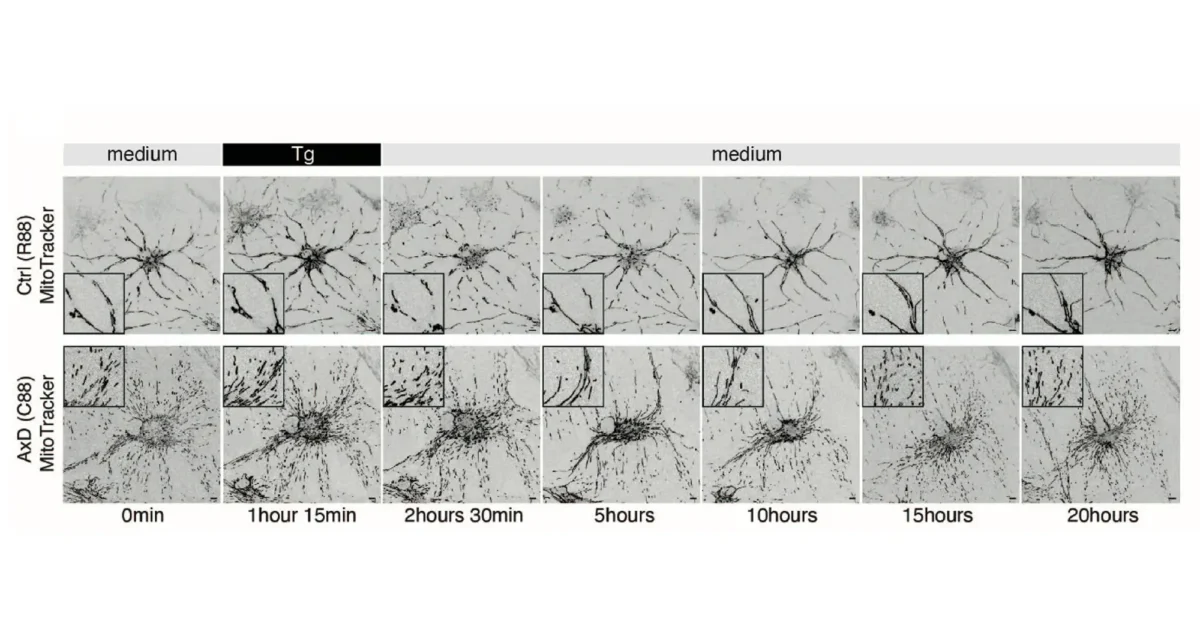
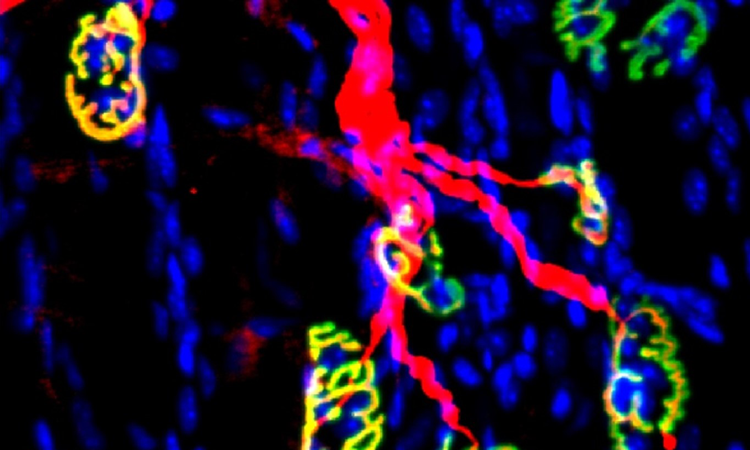
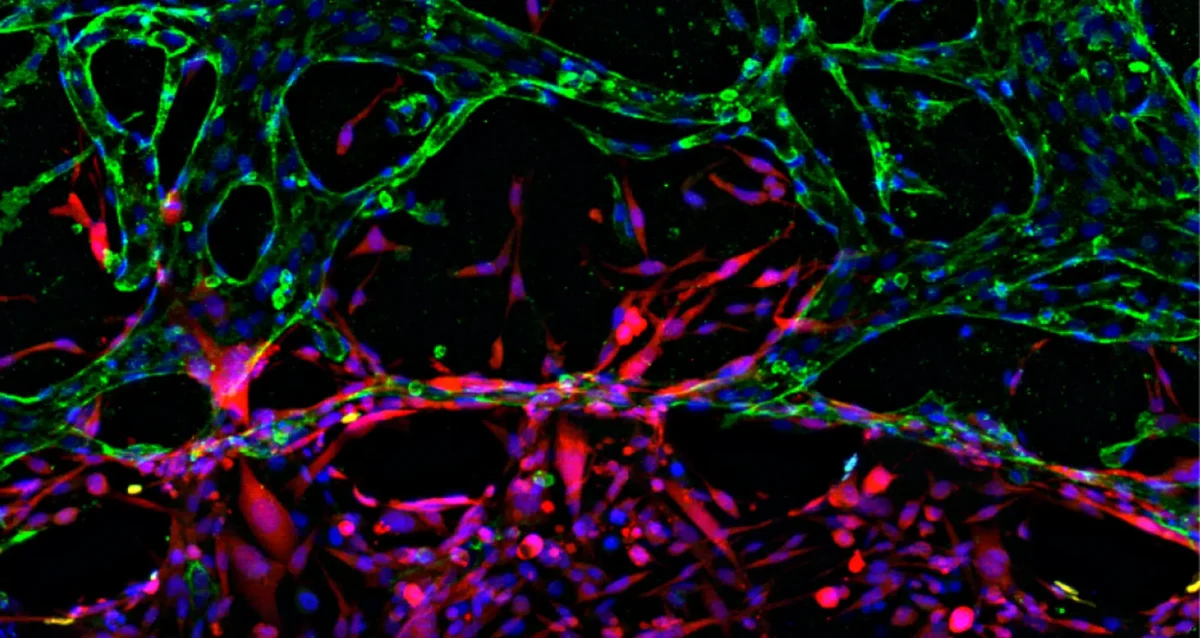
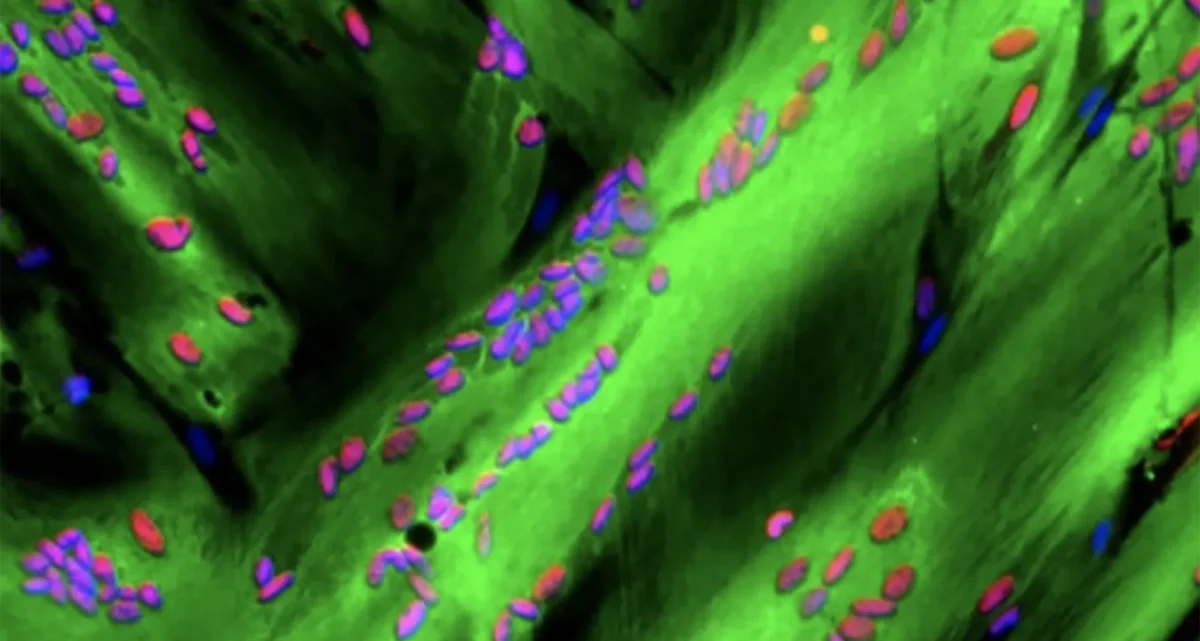
The Karin lab has established that the main type of primary liver cancer hepatocellular carcinoma (HCC) arises in the context of chronic liver injury and inflammation. To better understand this process we have developed methods for the isolation of HCC Progenitor Cell (HcPC) and have been studying how they are generated from normal, healthy, hepatocytes. This investigation has led us to make the surprising discovery that HcPC are generated from senescent hepatocytes, questioning whether cellular senescence is an effective tumor suppressive mechanism as was assumed till now. Curiously, in the context of MASLD and its more aggressive manifestation, MASH, metabolic liver diseases that affect 30% of Americans, hepatocyte senescence is triggered as a protective response that limits diet-induced DNA damage. Yet, by limiting hepatocyte DNA damage, senescence prevents the death of damaged hepatocytes keeping them as “ticking time bombs” that give rise to cancer once they have found ways to re-enter the cell cycle and start proliferating. Induction of hepatocyte senescence also seems to give rise to insulin resistance, and we are currently studying the mechanistic connection between the two, focusing on the role autocrine type I interferon signaling, which is activated in response to hepatocyte DNA damage.
Another, but less common, liver cancer that arises in chronically injured livers is intrahepatic cholangiocarcinoma (ICC). Although ICC is a cancer of the small intrahepatic bile ducts its cell of origin remains somewhat mysterious as it can either be the cells that form the bile ducts (cholangiocytes), hepatocytes or bipotential hepatobiliary progenitors. We have recently developed a new mouse model for a rare liver, inflammatory liver disease called Primary Sclerotizing Cholangitis (PSC) that shows progression to ICC and tumors with a mixed HCC-ICC phenotype. We are using this model to gain novel insights to the pathogenesis of both PSC and ICC.
MASLD/MASH, the two most common chronic liver diseases, which are caused by over-nutrition and obesity, are accompanied by substantial alterations to liver macrophage populations, leading to disappearance of resident liver macrophages or Kupffer Cells and the appearance of lipid associated macrophages (LAM), which are derived from recruited monocytes. We have recently found that LAMs are composed of two subclusters, pathogenic LAM-1 and protective LAM-2. We are investigating the diversification of LAM into LAM-1 and LAM-2 and investigating how LAM-2 exert their protective activity which leads to a marked reduction in MASH severity. We are also testing the therapeutic potential of LAM-2 transplantation.
Another inflammation promoted cancer is the most common pancreatic cancer -pancreatic ductal adenocarcinoma (PDAC). We have been studying how chronic inflammation and stress allow KRAS induced premalignant lesions progress to full blown invasive PDAC focusing on an EZH2-NRF2 controlled autoregulatory loop and activation of the collagen receptor DDR1, which is specifically engaged by a fragment of collagen I and other fibrillar collagens that is generated by matrix metalloproteases (MMPs). We are currently identifying the MMPs that are most critical for collagen cleavage in the context of PDAC and also investigating how the inhibition of collagen cleavage results in downregulation of DDR1 and interference with PDAC growth and progression. Moreover, we examining whether and how MASLD stimulates PDAC metastasis to liver.
 Jun 30, 2025
Jun 30, 2025Leading cancer and metabolic disease expert Michael Karin joins Sanford Burnham Prebys
Jun 30, 2025Karin will direct the Center for Metabolic and Liver Diseases and continue his studies on inflammation and cancer.
 May 19, 2016
May 19, 2016High levels of protein p62 predict liver cancer recurrence
May 19, 2016CANCER METABOLISM AND SIGNALING NETWORKS PROGRAM New research from SBP and UC San Diego shows that high levels of the…
 Feb 25, 2016
Feb 25, 2016Molecular “brake” prevents excessive inflammation
Feb 25, 2016Inflammation is a catch-22: the body needs it to eliminate invasive organisms and foreign irritants, but excessive inflammation can harm…