Researchers reveal a step-by-step guide to constructing stitched-together protein complexes capable of recycling most of the proteins in our cells
Scientists at Sanford Burnham Prebys have developed a clearer picture of how crucial machinery in the human cell’s recycling process for obsolete and misshapen proteins—known as proteasomes—are formed.
“Proteasomes are absolutely essential for health,” said Jianhua Zhao, PhD, an assistant professor in the Cancer Metabolism and Microenvironment Program and senior author on the new study. “An estimated 80% of proteins within the cell are degraded by proteasomes to maintain a healthy balance as new proteins replace those that have fulfilled their function or became misfolded and potentially harmful.”
Issues with proteasomes can contribute to certain cancers and are associated with age-related diseases, so a better understanding of how these prodigious protein degraders are made may lead to new treatments for many conditions.
Zhao and his team published results on September 18, 2024, in Nature Communications, that provide new details about stages in the process of proteasome assembly that had eluded investigators. Within a long and detailed molecular dance including more steps of choreography than a K-pop music video, earlier research attempts focused on the middle phases in the process.
“We have now provided data on the earlier and later steps of how proteasomes are constructed in cells,” said Hanxiao Zhang, a graduate student at Sanford Burnham Prebys and first author on the study. “This is really difficult to do as the later stages feature complex enzymatic reactions at a rapid pace, while the earlier steps focus on smaller and less stable protein complexes.”
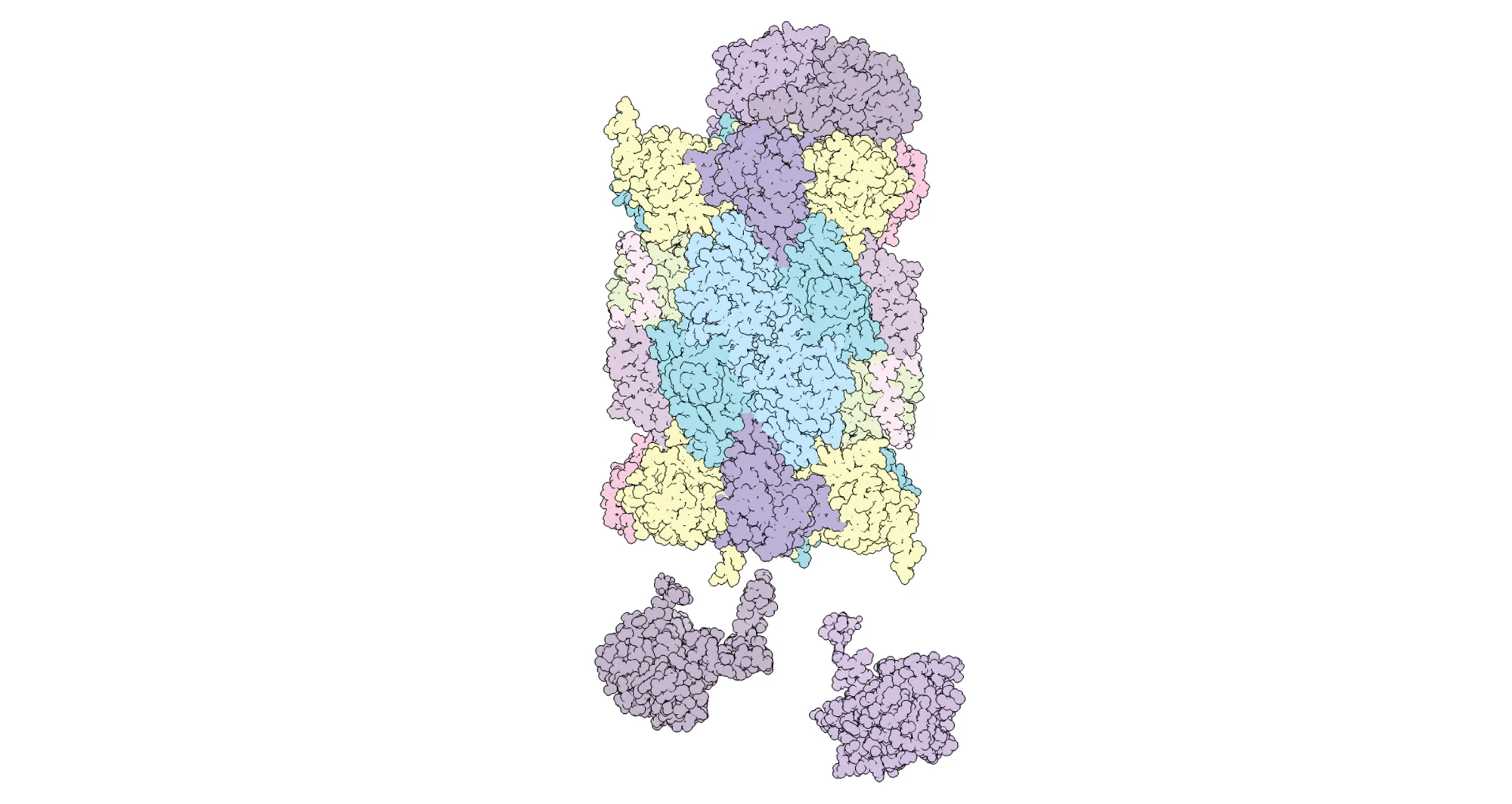
Zhang, Zhao and team investigated the fabrication of the canonical form of proteasome complexes found in humans, animals and plants. This proteasome, called the 20S proteasome, is marked by the coming together of four stacked rings comprised each of seven protein building blocks known as either alpha or beta subunits. The two rings of beta subunits at the center are sandwiched by two rings of alpha subunits.

Jianhua Zhao, PhD, is an assistant professor in the Cancer Metabolism and Microenvironment Program at Sanford Burnham Prebys.
The final 20S proteasome product looks less like cartoon mechanized lions forming a microscopic Voltron and more like a humble barrel, albeit just as destructive to proteins targeted for recycling. To see precisely how this barrel of proteins is joined, the researchers used gene editing techniques to tag helper proteins that bind with alpha and beta subunits and assist them in connecting to form their respective seven-piece rings.
To view the interactions between these marked faciliatory proteins—called chaperones—and the subunits as they link up into rings and stack in layers to form the 20S proteasome barrel, the scientists captured images using cryogenic electron microscopy (cryo-EM). This technology enables investigators to create 3D images of the cell and all its constituent parts that are accurate to the tiniest detail due to the ability to render individual atoms. Images taken using cryo-EM can be organized sequentially to develop films that show in real time how the cell’s many actors interact.
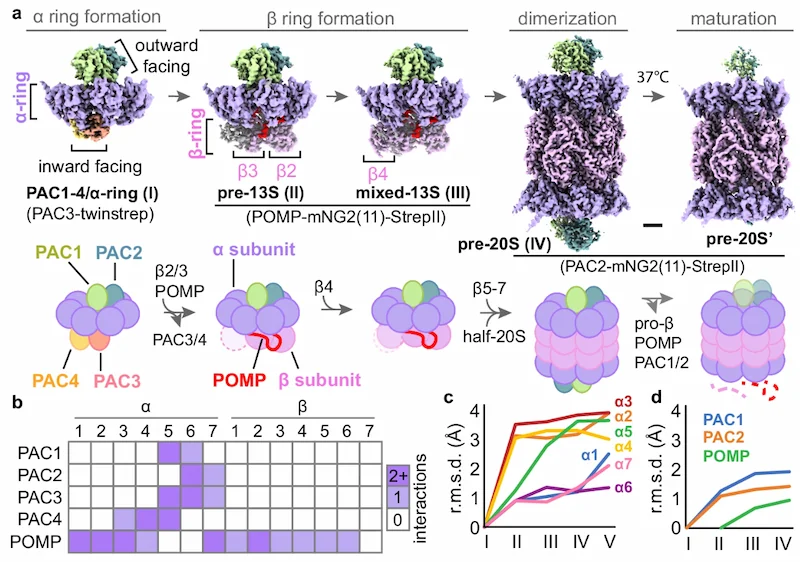
The first figure in the paper discusses several stages in the formation of the 20S proteasome, which is shaped like a barrel. This proteasome is marked by the coming together of four stacked rings comprised each of seven protein building blocks known as either alpha or beta subunits. The two rings of beta subunits at the center are sandwiched by two rings of alpha subunits.
The research team shed new light on how two protein chaperones bind on the top of the alpha subunit ring as it is constructed and are flanked by two chaperones on the bottom. With this initial ring taking the place of the metaphorical lid on what will become a barrel of four protein rings, the scientists also showed how the two bottom chaperones “under the lid” are removed and replaced by a different chaperone, and eventually by the rings of beta subunits that form the middle section of the 20S proteasome’s barrel shape.
“Learning more about the assembly of proteasomes may provide us with opportunities to leverage this knowledge to either target certain cancers or improve outcomes with aging,” said Zhao. “We also plan to use this data to make our own proteasomes in the laboratory and potentially design better proteasomes.”
Zhao believes that this may enable the findings to have wider implications beyond cancer and aging.
“What we’re looking at is not necessarily something that’s limited to humans,” added Zhao. “We’re also looking at host and pathogen interactions.”
This potential avenue led Zhao to collaborate with Anthony O’Donoghue, PhD, an associate professor in the Skaggs School of Pharmacy and Pharmaceutical Sciences at the University of California San Diego, to develop proteasome-based inhibitors to treat parasitic infections.
Additional authors on the study include Chenyu Zhou and Zarith Mohammad.
The study was supported by the National Institutes of Health (R35 GM147487 and S10 OD026926) and National Cancer Institute (Cancer Center Support Grant P30 CA030199).
Molecular graphics and analyses were performed with UCSFChimeraX, which is supported by the NIH (R01-GM129325) and the National Institute of Allergy and Infectious Diseases Office of Cyber Infrastructure and Computational Biology.
The study’s DOI is 10.1038/s41467-024-52513-0.