Scientists create a machine-learning algorithm that automates high-throughput screens of epigenetic medicines.
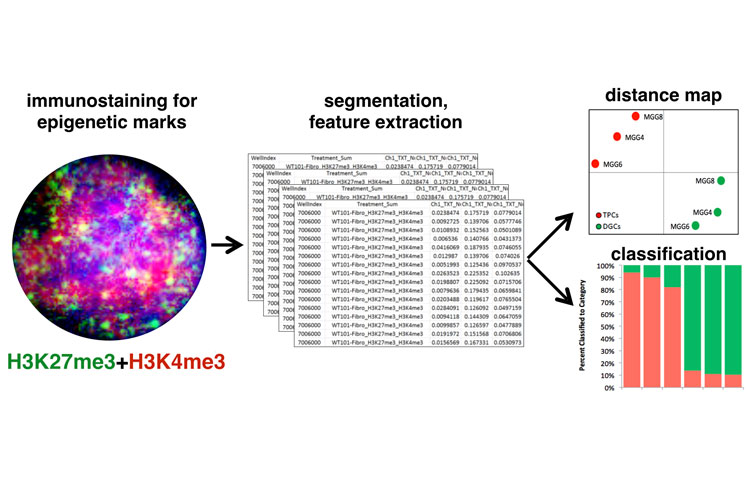
Machine learning’s powerful ability to detect patterns in complex data is revolutionizing how we drive, how we diagnose disease and now, how we discover new drugs. Scientists at Sanford Burnham Prebys Medical Discovery Institute have developed a machine-learning algorithm that gleans information from microscope images—allowing for high-throughput epigenetic drug screens that could unlock new treatments for cancer, heart disease, mental illness and more. The study was published in eLife.
“In order to identify the rare few drug candidates that induce desired epigenetic effects, scientists need methods to screen hundreds of thousands of potential compounds,” says Alexey Terskikh, PhD, associate professor in Sanford Burnham Prebys’ Development, Aging and Regeneration Program and senior author of the study. “Our study describes a powerful image-based approach that enables high-throughput epigenetic drug discovery.”
Epigenetics refers to chemical tags on DNA that allow cellular machinery greater or less access to genes—thus altering gene expression. Nearly all changes in a cell, including reaction to a drug and environmental stress, are reflected by its epigenetic state. Several medicines that target epigenetic alterations are approved by the U.S. Food and Drug Administration (FDA) for the treatment of cancer, and researchers are working to find additional epigenetic-based therapies. However, drug discovery has been slowed by a lack of a high-throughput screening method: Scientists currently visualize epigenetic changes using special dyes and traditional microscopy methods.
“Epigenetic changes contribute to or are at the root of numerous neurological disorders, including brain tumors and some forms of autism,” says Harley Kornblum, MD, PhD, professor and associate director of the Intellectual and Developmental Disabilities Research Center in the Semel Institute for Neuroscience and Human Behavior at the David Geffen School of Medicine at UCLA. “This method will allow for the rapid assessment of the epigenetic state of individual cells and their response to potential treatments. I am eager to apply it to my own laboratory’s work to understand and treat these disorders.”
In the study, the scientists trained a machine-learning algorithm using a set of more than 220 drugs known to work epigenetically. The resulting method, called Microscopic Imaging of Epigenetic Landscapes (MIEL), was able to detect active drugs, classify the compounds by their molecular function, spot epigenetic changes across multiple cell lines and drug concentrations, and help identify how unknown compounds work. The scientists used the approach to identify epigenetic compounds that may be able to help treat glioblastoma, a deadly brain cancer.
“Our method is ready for immediate use by pharmaceutical companies looking to develop epigenetic drug screens,” says Chen Farhy, PhD, a postdoctoral researcher in the Terskikh lab and first author of the study. “Industry and academic researchers working on mechanistic studies may also benefit from this method, as the algorithm can detect and categorize epigenetic changes induced by experimental treatments, genetic manipulations or other approaches.”
Terskikh and his team are already using the algorithm to study epigenetic changes in aging cells, with the aim of developing compounds that promote healthy aging—the single greatest risk factor for disease. This work is conducted in collaboration with Sanford Burnham Prebys professor Peter Adams, PhD Terskikh is also eager to broaden the technology from 2D images to 3D videos, which will expand the power of the approach.
Additional study authors include Luis Orozco, Fu-Yue Zeng, PhD, Ian Pass, PhD, and Chun-Teng Huang of Sanford Burnham Prebys; Jarkko Ylanko and Santosh Hariharan of Sunnybrook Research Institute; Fernando Ugarte, PhD, and Camilla Forsberg, PhD, of the University of California, Santa Cruz; and David Andrews, PhD, of Sunnybrook Research Institute and University of Toronto. Contact Terskikh or Farhy for more information about using the algorithm in your research.
Research reported in this press release was supported by Celgene, the U.S. National Institutes of Health (NIH) (R01NS066278), the Canadian Institutes of Health Research (FDN143312) and a Tier 1 Canada Research Chair in Membrane Biogenesis award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The study’s DOI is 10.7554/eLife.49683.