Rapidly evolving computational tools may unlock vast archives of untapped clinical information—and help solve complex challenges confronting healthcare providers
The wealth of data stored in electronic medical records has long been considered a veritable treasure trove for scientists able to properly plumb its depths.
Emerging computational techniques and data management technologies are making this more possible, while also addressing complicated clinical research challenges, such as optimizing the design of clinical trials and quickly matching eligible patients most likely to benefit.
Scientists are also using new methods to find meaning in previously published studies and creating even larger, more accessible datasets.
“While we are deep in the hype cycle of artificial intelligence [AI] right now, the more important topic is data,” says Sanju Sinha, PhD, an assistant professor in the Cancer Molecular Therapeutics Program at Sanford Burnham Prebys. “Integrating data together in a clear, structured format and making it accessible to everyone is crucial to new discoveries in basic and clinical biomedical research.”
Sinha is referring to resources such as the St. Jude-Washington University Pediatric Cancer Genome Project, which makes available to scientists whole genome sequencing data from cancerous and normal cells for more than 800 patients.
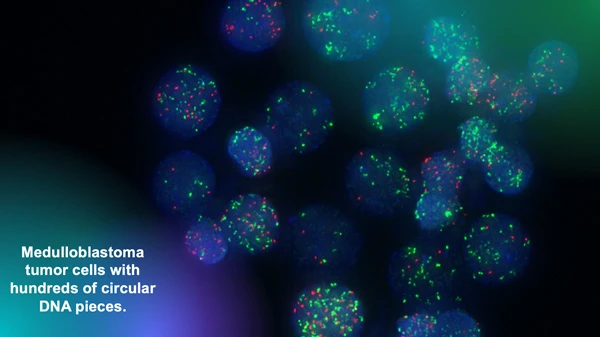
The Chavez lab uses fluorescent markers to observe circular extra-chromosomal DNA elements floating in cancer cells. Research has shown that these fragments of DNA are abundant in solid pediatric tumors and associated with poor clinical outcomes. Image courtesy of Lukas Chavez.
The Children’s Brain Tumor Network is another important repository for researchers studying pediatric brain cancer, such as Lukas Chavez, PhD, an assistant professor in the Cancer Genome and Epigenetics Program at Sanford Burnham Prebys.
“We have analyzed thousands of whole genome sequencing datasets that we were able to access in these invaluable collections and have identified all kinds of structural rearrangements and mutations,” says Chavez. “Our focus is on a very specific type of structural rearrangement called circular extra-chromosomal DNA elements.”
Circular extra-chromosomal DNA elements (ecDNA) are pieces of DNA that have broken off normal chromosomes and then been stitched together by DNA repair mechanisms. This phenomenon leads to circular DNA elements floating around in a cancer cell.

Sanju Sinha, PhD, is an assistant professor in the Cancer Molecular
Therapeutics Program at Sanford Burnham Prebys.
“We have shown that they are much more abundant in solid pediatric tumors than we previously thought,” adds Chavez. “And we have also shown that they are associated with very poor outcomes.”
To help translate this discovery for clinicians and their patients, Chavez is testing the use of deep learning AI algorithms to identify tumors with ecDNA by analyzing the biopsy slides that are routinely created by pathologists to diagnose brain cancer.
“We have already done the genomic analysis, and we are now turning our attention to the histopathological images to see how much of the genomic information can be predicted from these images,” says Chavez. “Our hope is that we can identify tumors that have ecDNA by evaluating the images without having to go through the genomic sequencing process.”
Currently, this approach serves only as a clinical biomarker of a challenging prognosis, but Chavez believes it can also be a diagnostic tool—and a game changer for patients.
“I’m optimistic that in the future we will have drugs that target these DNA circles and improve the therapeutic outcome of patients,” says Chavez.
“Once medicine catches up, we need to be able to find the patients and match them to the right medicine,” says Chavez. “We’re not there yet, but that’s the goal.”
Chavez is also advancing his work as scientific director of the Pediatric Neuro-Oncology Molecular Tumor Board at Rady Children’s Hospital in San Diego.
“Recently, it has been shown that new sequencing technologies coupled with machine learning tools make it possible to compress the time it takes to sequence and classify types of tumors from days or weeks to about 70 minutes,” says Chavez. “This is quick enough to take that technology into the operating room and use a surgical biopsy to classify a tumor.
“Then we could get feedback to the surgeon in real time so that more or less tissue can be removed depending on if it is a high- or low-grade tumor—and this could dramatically affect patient outcomes.
“When I talk to neurosurgeons, they are always in a pickle between trying to be aggressive to reduce recurrence risk or being conservative to preserve as much cognitive function and memory as possible for these patients.
“If the surgeon knows during surgery that it’s a tumor type that’s resistant to treatment versus one that responds very favorably to chemotherapy, radiation or other therapies, that will help in determining how to strike that surgical balance.”

Lukas Chavez, PhD, is an assistant professor in the Cancer Genome and Epigenetics Program at Sanford Burnham Prebys.
Artist’s rendering of X-shaped chromosomes floating in a cell alongside circular extra-chromosomal DNA elements.
Rady Children’s Hospital has also contributed to the future of genomic and computational medicine through BeginNGS, a pilot project to complement traditional newborn health screening with genomic sequencing that screens for approximately 400 genetic conditions.
“The idea is that if there is a newborn baby with a rare disease, their family often faces a very long odyssey before ever reaching a diagnosis,” says Chavez. “By sequencing newborns, this program has generated success stories, such as identifying genetic variants that have allowed the placement of a child on a specific diet to treat a metabolic disorder, and a child to receive a gene therapy to restore a functional immune system.”
Programming in a Petri Dish, an 8-part series
How artificial intelligence, machine learning and emerging computational technologies are changing biomedical research and the future of health care
- Part 1 – Using machines to personalize patient care. Artificial intelligence and other computational techniques are aiding scientists and physicians in their quest to prescribe or create treatments for individuals rather than populations.
- Part 2 – Objective omics. Although the hypothesis is a core concept in science, unbiased omics methods may reduce attachments to incorrect hypotheses that can reduce impartiality and slow progress.
- Part 3 – Coding clinic. Rapidly evolving computational tools may unlock vast archives of untapped clinical information—and help solve complex challenges confronting health care providers.
- Part 4 – Scripting their own futures. At Sanford Burnham Prebys Graduate School of Biomedical Sciences, students embrace computational methods to enhance their research careers.
- Part 5 – Dodging AI and computational biology dangers. Sanford Burnham Prebys scientists say that understanding the potential pitfalls of using AI and other computational tools to guide biomedical research helps maximize benefits while minimizing concerns.
- Part 6 – Mapping the human body to better treat disease. Scientists synthesize supersized sets of biological and clinical data to make discoveries and find promising treatments.
- Part 7 – Simulating science or science fiction? By harnessing artificial intelligence and modern computing, scientists are simulating more complex biological, clinical and public health phenomena to accelerate discovery.
- Part 8 – Acceleration by automation. Increases in the scale and pace of research and drug discovery are being made possible by robotic automation of time-consuming tasks that must be repeated with exhaustive exactness.