- Google Scholar Kevin Yip Google Scholar Profile
- H1 Connect F1000 Kevin Yip Faculty Profile
- Bioinformatics micro-modules Watch | Kevin Yip’s Channel for Learning Bioinformatics Concepts, Algorithms and Data Structures
Phenomena or Process: Transcriptional Regulation
Dr. Yu Xin (Will) Wang received his PhD at the University of Ottawa where he identified cellular asymmetry and polarity mechanisms regulating muscle stem cell self-renewal and skeletal muscle regeneration. He then carried out postdoctoral training at Stanford University School of Medicine developing single cell multi-omic approaches to characterize the regenerative process and what goes awry with disease and aging.
“I’ve always had a passion for science and became fascinated with how the body repairs and heals itself when I was introduced to the potential of stem cells in regenerative medicine. I was struck by the ability of a small pool of muscle stem cells that can rebuild and restore the function of muscle. My lab at Sanford Burnham Prebys aims to better understanding the repair process and harness our body’s ability to heal in order to combat chronic diseases and even counteract aging.“
Education and Training
Postdoctoral Fellowship, Stanford University School of Medicine
PhD in Cellular Molecular Medicine, University of Ottawa, Canada
BS in Biomedical Sciences, University of Ottawa, Canada
Prestigious Funding Awards
2020: NINDS K99/R00 Pathway to Independence Award
Honors and Recognition
Governor General’s Gold Medal – Canada
Related Disease
Aging-Related Diseases, Amyotrophic Lateral Sclerosis (Lou Gehrig’s Disease), Arthritis, Cachexia, Inflammatory/Autoimmune Disease, Multiple Sclerosis, Muscular Dystrophy, Myopathy, Neurodegenerative and Neuromuscular Diseases, Sarcopenia/Aging-Related Muscle Atrophy, Spinal Muscular Atrophy
Phenomena or Processes
Adult/Multipotent Stem Cells, Aging, Cell Signaling, Development and Differentiation, Epigenetics, Exercise, Extracellular Matrix, Neurogenesis, Organogenesis, Regenerative Biology, Transcriptional Regulation
Anatomical Systems and Sites
Immune System and Inflammation, Musculoskeletal System, Nervous System
Research Models
Clinical and Transitional Research, Computational Modeling, Human Adult/Somatic Stem Cells, Mouse
Techniques and Technologies
3D Image Analysis, Bioinformatics, Cellular and Molecular Imaging, Gene Knockout (Complete and Conditional), Genomics, High Content Imaging, High-Throughput/Robotic Screening, Live Cell Imaging, Machine Learning, Microscopy and Imaging, Proteomics, Transplantation
The Wang lab is interested in elucidating critical cell-cell interactions that mediate the function of tissue-specific stem cells during regeneration and disease, with a focus on how a coordinated immune response can promote regeneration and how autoimmunity impacts tissue function and hinder repair.
Specifically, the Wang lab aims to identify cellular and molecular crosstalk between muscle, nerve, and immune systems to develop targeted therapies that overcome autoimmune neuromuscular disorders and autoimmune aspects of “inflammaging.”
Yu Xin (Will) Wang’s Research Report
The lab’s research is translationally oriented and utilizes interdisciplinary molecular, genetic, computational (machine learning and neural networks), and bioengineering approaches to view biology and disease from new perspectives. We combine multi-omics sequencing and imaging methods to resolve how different cell types work together after injury to repair tissues and restore function. We use a data-driven approach to identify targetable disease mechanisms and, through collaborations with other researchers and clinicians, develop therapies that promote regeneration. Visit our lab website to learn more.
 Jun 12, 2025
Jun 12, 2025Turning back time on muscle stem cells to prevent frailty from aging
Jun 12, 2025Study from Dr. Will Wang’s lab finds a way to restore stem cells from aged muscle to become young again…
 Aug 20, 2024
Aug 20, 2024Mapping the human body to better treat disease
Aug 20, 2024Scientists are investigating the inner workings of our bodies and the cells within them at an unprecedented level of detail.
 Aug 13, 2024
Aug 13, 2024Dodging AI and other computational biology dangers
Aug 13, 2024Sanford Burnham Prebys scientists say that understanding the potential pitfalls of using artificial intelligence and computational biology techniques in biomedical…
 Oct 11, 2023
Oct 11, 2023Inhibiting an enzyme associated with aging could help damaged nerves regrow and restore strength
Oct 11, 2023New research has demonstrated a way to accelerate recovery from peripheral nerve injury by targeting an enzyme that was thought…
 Jan 26, 2023
Jan 26, 2023Three big questions for cutting-edge biologist Will Wang
Jan 26, 2023Will Wang’s spatial omics approach to studying neuromuscular diseases is unique.
 Nov 23, 2022
Nov 23, 2022Yu Xin (Will) Wang joins Sanford Burnham Prebys to advance regenerative medicine
Nov 23, 2022Molecular biologist Yu Xin (Will) Wang, PhD, has joined Sanford Burnham Prebys as an assistant professor in the Development, Aging,…
Xueqin (Sherine) Sun seeks to better understand the genetic and epigenetic underpinnings of cancers, using genome editing technologies, animal and patient-derived models, and other tools to develop more effective cancer therapies.
“My lab is interested in studying how DNA or the machinery that interprets it leads to the transformation of normal cells into cancerous cells and concurrently, their specific vulnerabilities. Identifying these intrinsic vulnerabilities and targeting them properly is profoundly important to developing effective cancer therapies.”
Another aspect of Sun’s work is understanding how cancer cells and tumors change their circumstances and environment to improve survival, including hiding from or repressing the immune system.
“Changes to DNA itself and the way how DNA is interpreted by cells can transform normal cells into cancer cells. And transformed cells propagate by enhancing the misinterpreted DNA information, which in turn becomes the Achilles’ heel of cancer cells. Our goal is to find out how DNA information is misinterpreted in different ways and how to correct it to halt cancer.”
At Sanford Burnham Prebys, Sun and colleagues will employ a host of leading-edge tools and approaches, including functional genomics, artificial intelligence, structural biology, large-scale drug screening, and advanced imaging/spatial technologies.
Sun conducted her postdoctoral fellowship at Cold Spring Harbor Laboratory under the guidance of Alea Mills, PhD, a professor at the National Cancer Institute-designated cancer center at Cold Spring Harbor.
She received her PhD from Wuhan University in China.
Related Disease
Aging-Related Diseases, Brain Cancer, Cancer, Childhood Diseases, Immune Disorders, Inflammatory/Autoimmune Disease, Leukemia/Lymphoma
Phenomena or Processes
Adapter Proteins, Adult/Multipotent Stem Cells, Aging, Angiogenesis, Apoptosis and Cell Death, Bcl-2 Family, Cancer Biology, Cancer Epigenetics, Cell Adhesion and Migration, Cell Biology, Cell Cycle Progression, Cell Differentiation, Cell Motility, Cell Proliferation, Cell Signaling, Cell Surface Receptors, Cellular Senescence, Chromosome Dynamics, Combinatorial Therapies, Cytokines, Development and Differentiation, Disease Therapies, DNA Damage Checkpoint Function, Embryonic/Pluripotent Stem Cells, Epigenetics, Gene Regulation, Genomic Instability, Growth Factors, Hematopoiesis, Host Defense, Host-Pathogen Interactions, Inflammation, Innate Immunity, Kinase Inhibitors, Metastasis, Neurogenesis, Oncogenes, Phosphorylation, Posttranslational Modification, Receptor Tyrosine Kinases, Serine/Threonine Kinases, Signal Transduction, TNF-Family, Transcription Factors, Transcriptional Regulation, Tumor Microenvironment, Tumorigenesis, Tyrosine Kinases, Ubiquitin, Ubiquitin Protease System and Ubiquitin-like Proteins
Anatomical Systems and Sites
Brain, General Cell Biology, Hematopoietic System, Immune System and Inflammation, Nervous System
Research Models
Bacteria, Cultured Cell Lines, Human Adult/Somatic Stem Cells, Human Cell Lines, Mouse, Mouse Cell Lines, Mouse Embryonic Stem Cells, Mouse Somatic Stem Cells, Primary Cells, Primary Human Cells
Techniques and Technologies
3D Image Analysis, 3D Reconstructions, Biochemistry, Bioinformatics, Cell Biology, Cellular and Molecular Imaging, Chemical Biology, Computational Biology, Confocal Microscopy, Correlative Light and Electron Microscopy, Drug Delivery, Drug Discovery, Drug Efficacy, Electron Microscopy, Fluorescence Microscopy, Fragment-Based Drug Design, Gene Expression, Gene Knockout (Complete and Conditional), Gene Silencing, Genetics, Genomics, High Content Imaging, High-Throughput/Robotic Screening, In vivo Modeling, Live Cell Imaging, Live Imaging, Mass Spectrometry, Microscopy and Imaging, Molecular Biology, Molecular Genetics, Nucleic Acid Synthesis, Protein-Protein Interactions, Protein-Small Molecule Interactions, Proteomics, Rational Drug Design, RNA Interference (RNAi), Scanning Cytometry, Small Molecule Compounds, Transgenic Organisms, Transplantation
We seek to understand why cancer occurs and what is the Achille’s heel of cancer, and to develop effective therapeutic interventions.
The successful treatment of any disease requires a good understanding of the mechanisms at work. Cancer is fundamentally caused by aberrant gene expression, which reflects the misinterpretation of DNA information at both genetic and epigenetic levels. We are interested in uncovering DNA-related alterations that drive cancer-favored transcriptional programs, identifying cancer-specific vulnerabilities, and developing effective therapeutic interventions for cancer treatment.
Xueqin Sun’s Research Report
Precise gene expression (the interpretation of DNA) is essential for almost all biological processes, and understanding gene regulation is one of the most pivotal frontiers in biological research under both health and disease circumstances. Gene expression is mainly regulated at genetic (with changes of DNA sequence) and epigenetic (without changing DNA sequence) levels. And gene dysregulation can lead to various health conditions and diseases, including developmental disorders, aging, and cancer. The overarching goal of Sun Lab is to uncover driving genetic and epigenetic alterations involved in cancer, to understand how developmental pathways and aging process impact cancer progression, and to identify mechanisms of action for developing more effective therapeutic strategies.
We are an interdisciplinary lab particularly focused on the following research directions:
- The EP400 chromatin remodeling complex
The EP400 complex is an evolutionarily conserved SWR1-class ATP-dependent chromatin remodeling complex encompassing ~17 components, with a total molecular mass of ~1.5 mega-dalton. The EP400 complex plays critical roles in diverse cellular processes, including chromosome stability, transcription, DNA recombination, DNA damage repair, embryonic stem cell renewal/development, and oncogenesis. The EP400 complex can incorporate histone variants, such as H2AZ and H3.3, into the genome to regulate gene expression. Our recent work discovers BRD8—one of the core subunits of the EP400 complex—as a unique vulnerability of p53 wildtype glioblastoma (GBM), the most prevalent and devastating type of brain cancer. BRD8-driven EP400 complex highjacks H2AZ at p53 target loci to block p53-mediated transactivation and tumor suppression (Nature, 2023). The bromodomain of BRD8 plays the key role in this process. Bromodomain is a druggable domain as evidenced by a number of successful small molecules targeting diverse bromodomains encoded by the human genome across multiple cancer types. Furthermore, findings from others and us suggest that the EP400 complex is involved in different cancers. Thus, we seek to unravel the roles of the EP400 complex in health and disease, and to better understand how to target the EP400 complex for developing effective therapeutic interventions. - The NuRD chromatin remodeling complex
The NuRD complex is also a highly conserved class of ~ 1 MDa multi-subunit chromatin remodeling complexes that consume energy derived from ATP hydrolysis to remodel the configuration of chromatin to control gene transcription programs, with a primary role in gene silencing. Chromatin remodeling is vital for efficiently framing the cellular response to both intrinsic and extrinsic signals and has enormous implications for determining cellular states. NuRD complex is unique in combining ATP-dependent chromatin remodeling, protein deacetylase activity, and recognition of methylated DNA and histone modifications, and has multifarious roles in chromatin organization, transcription regulation, and genome maintenance; thereby, largely impacts health and disease. The NuRD complex has been in the central stage of brain development studies, and is significantly related to brain disorders/diseases. Interestingly, NuRD complex re-assembles by exchanging the chromatin remodeling subunits CHD3/4/5 to achieve specific regulation of an array of genes required for generating distinct cell types in a highly organized manner, especially over brain development. Amongst the genes encoding NuRD complex components, CHD5 is located in human chromosome 1 short arm (1p36), a region that is frequently hemizygously deleted in diverse cancers. Besides genetic deletion, CHD5 is also often silenced in cancer cells due to epigenetic mechanisms, such as promoter hypermethylation, aberrant expression of other chromatin regulators, and microRNAs-mediated translational repression and/or mRNA instability. Our current work seeks to determine whether and how CHD5-driven NuRD complex is involved in tumorigenesis (In preparation, 2024). We will further understand how NuRD complex is involved in both development and tumorigenesis, and identify mechanism of action to develop rational therapeutic strategies. - Novel genetic and epigenetic underpinnings in GBM
GBM is notorious for being a highly complex and plastic cancer type. However, at the genetic level, GBM harbors a relatively low genetic alteration burden compared to the majority of other cancers from pan-cancer profiling studies. This indicates the largely undocumented epigenetic mechanisms that interplay with genetic alterations and co-reprogram transcriptional networks essential for GBM development. Epigenetic changes are usually reversible by nature, as evidenced by numerous successes in targeting epigenetic regulators using small chemical compounds. As actionable therapeutic targets for GBM have been scarce, we are keen to uncover novel epigenetic pathways underlying gliomagenesis under different genetic backgrounds, which will potentially provide promising therapeutic opportunities for GBM treatment. - Novel GBM mouse models
Despite decades of effort, our knowledge about GBM biology is still very limited. GBM harbors a number of genetic alterations. However, among these recurrent genetic lesions, only several have been implicated in gliomagenesis, with most being undocumented. Moreover, the mechanisms by which these genetic alterations are involved in establishing GBM-favored epigenetic landscapes and transcription programs during GBM progression are still largely elusive. The lack of efficient approach to establish mouse models for investigating gene function in gliomagenesis and the limit of current mouse models to recapitulate clinical GBM features in brain is the prime reason that hinders GBM biological research. To this end, we have developed an engineered neural stem cells (NSCs)-based strategy to rapidly generate highly aggressive GBM with desired genetic lesions (genotypes) in mouse brain. Therefore, we will further optimize this strategy to establish a series of novel mouse models possessing recurrent combinations of genetic alterations (genotypes) in GBM, in order to systematically study whether and how these genetic lesions are involved in gliomagenesis and identify genotype-specific dependencies. - Crosstalk between GBM cells and tumor microenvironment
GBM exhibits highly diffuse and infiltrative nature, which contributes to therapeutic resistance and tumor relapse after surgical removal, resulting in dismal prognosis. A better understanding of gliomagenesis involving not only malignant cells themselves, but also the holistic bidirectional interactions of malignant cells with a variety of proximal and distal cells within the organism, is profound for developing novel effective therapies to improve GBM prognosis. Individual invasive GBM cells intermingle with normal brain cells and often cause relapse in brain areas essential for patient survival. Emerging evidence indicates that glioma cells highjack normal brain cells to thrive, and even transform them. However, how gliomagenesis reshapes ecological composition/landscape in host brain and how brain microenvironment affects gliomagenesis are still largely unclear. By using our novel highly invasive mouse models that recapitulate the multiforme diffuse topographies of GBM in brain, we seek to understand the interactions between GBM cells and brain microenvironment, and identify extrinsic pathways that are essential for GBM progression and migration.
Our lab is focused on both fundamental questions in cancer biology and translation of promising therapeutic strategies.
To achieve these, we work together with many fantastic collaborators to develop and leverage cutting-edge technologies, including but not limited to, high-throughput functional genomics (CRISPR/Cas9 screens, exon tiling scan, targeted mutagenesis, etc.), cell and molecular biology, genomics, epigenomics, proteomics, biochemistry, microscopy (2D/3D, time-lapse, two-photon, light sheet, etc.), automated large-scale drug synthesis/screening, structural biology, single cell and spatial multi-omics, artificial intelligence, and bioinformatics. We also establish novel patient-derived models and novel mouse models to facilitate our research programs. Our ultimate goals are to better understand fundamental genetic and epigenetic apparatuses involved in cancer-specific transcriptional networks, provide more effective therapeutic opportunities, and contribute to shifting the paradigms in cancer treatment and precision medicine.
 Aug 19, 2024
Aug 19, 2024Women in Science event at Sanford Burnham Prebys examines how female faculty members navigate research careers
Aug 19, 2024Topics at the event included work/life balance, caregiving and family obligations, and gender disparities in academic rank at research and…
 Mar 13, 2024
Mar 13, 2024Xueqin Sun seeks to illuminate the underlying causes of cancer
Mar 13, 2024New Sanford Burnham Prebys scientist investigates the mutational powers of cancer cells — and their vulnerabilities
 Dec 19, 2023
Dec 19, 2023Sanford Burnham Prebys continues unprecedented recruitment of early-career scientists
Dec 19, 2023Continuing its rapid and dramatic recruitment of emerging, top-tier researchers, Sanford Burnham Prebys has hired two more highly regarded early-career…
Pier Lorenzo Puri earned his MD at the University of Rome “la Sapienza” in 1991. Dr. Puri completed his internship in Internal Medicine at the hospital “Policlinico Umberto I” (Rome) from 1992 to 1997, and defended an experimental thesis on the vascular effects of angiotensin II to graduate as Specialist in Internal medicine at the University of Rome “la Sapienza” in 1997. During this time he was frequently working at the Freien University of Berlin, as visiting scientist at the Deprtment of Biochemistry and Molecular Biology, to perform experiments of protein and DNA microinjection in cultured cells. Dr. Puri trained as a post-doctoral fellow at the University of California San Diego (UCSD), in the department of Cell Biology, under the supervision of Dr. Wang, from 1997 to 2001. He was appointed as Staff Scientist at the Salk Institute (La Jolla) in 2001, and became an Assistant Telethon Scientist at the Dulbecco Telethon Institute in Rome in 2002. He was upgraded to Associate Telethon Scientist at the Dulbecco Telethon Institute in Rome since 2007 and became Senior Telethon Scientist, Dulbecco Telethon Institute, in 2012, but declined this position. Dr. Puri joined Sanford Burnham Prebys as an Assistant Professor in 2004. He has been promoted to Associate Professor in 2010 and full Professor in 2015. From 2008 to 2016 Dr. Puri served as Adjunct Professor of Pediatrics at the University of California, San Diego. From 2008 to 2013 Dr Puri was an Associate Member of Sanford Children’s Health Research Center. Dr Puri has been Director of the laboratory of Epigenetics and Regeneration at Fondazione S. Lucia, Roma, Italy, but stepped down this position since 2019.
Education
University of California San Diego, Postdoctoral, Department of Biology
University of Rome La Sapienza, PhD, Internal Medicine
University of Rome La Sapienza, MD, Internal Medicine
University of Rome La Sapienza, Undergraduate, Internal Medicine
Other Appointments
2020-2024: Member of the Science Advisory Board (SAB) European Commission-funded Consortium BIND (Brain Involvement In Dystrophinopathies)
2015-2019: Standing Member, NIH Study Section (SMEP)
2010-present: Member of Editorial Board of Skeletal Muscle
Related Disease
Aging-Related Diseases, Childhood Diseases, Molecular Biology, Muscular Dystrophy, Myopathy, Neurodegenerative and Neuromuscular Diseases, Rhabdomyosarcomas, Sarcopenia/Aging-Related Muscle Atrophy, Spinal Cord Injury, Transcription Factors
Phenomena or Processes
Adult/Multipotent Stem Cells, Aging, Cell Biology, Cell Cycle Progression, Cell Differentiation, Cell Signaling, Cellular Senescence, Development and Differentiation, Disease Therapies, DNA Damage Checkpoint Function, Epigenetics, Gene Regulation, Phosphorylation, Regenerative Biology, Signal Transduction, Transcriptional Regulation
Anatomical Systems and Sites
General Cell Biology, Musculoskeletal System
Research Models
Clinical and Transitional Research, Cultured Cell Lines, Human Adult/Somatic Stem Cells, Mouse Embryonic Stem Cells, Mouse Somatic Stem Cells, Primary Human Cells
Techniques and Technologies
Bioinformatics, Cellular and Molecular Imaging, Gene Expression, Genomics
Puri’s lab group investigates the molecular and epigenetic regulation of gene expression in skeletal muscle progenitors and other muscle-resident cell types (including fibro-adipogenic progenitors, cells from the inflammatory infiltrate, cellular components of neuro-muscular junctions) during physiological and pathological perturbations of skeletal muscle homeostasis.
- We use molecular, biochemical and epigenetic tools to understand structural and functional principles of the 3D genome organization that regulates gene expression during muscle regeneration and diseases.
- A topic of particular interest is the analysis of chromatin interactions that define the 3D genome organization and the identification of structural and functional interactions that regulate cell type-specific patterns of gene expression in response to cues released within the skeletal muscle regenerative environment in health and disease conditions, such as muscular dystrophies and other neuromuscular diseases.
- The knowledge derived from our studies is instrumental to elucidate the pathogenesis of muscular disorders and discover pharmacological interventions that promote muscle regeneration to repair diseased muscles.
- Current translational focus is devoted to:
- the study of the therapeutic potential of HDAC inhibitors for treatment of Duchenne Muscular Dystrophy (DMD)
- the identification of genome variants associated to DMD patient-specific patterns of expression of disease-modifier genes that can account for individual trends of disease progression beyond the common genetic deficiency of dystrophin
- the effect of dystrophin deficiency and restoration by gene therapy on 3D genome and transcriptional output of DMD myofibers; the therapeutic potential of extracellular vesicles released by fibro-adipogenic progenitors of DMD skeletal muscles exposed to HDACi.

Puri Lab
Pier Lorenzo Puri’s Research Report
Puri’s lab group investigates the molecular and epigenetic regulation of gene expression in skeletal muscle progenitors and other muscle-resident cell types (including fibro-adipogenic progenitors, cells from the inflammatory infiltrate, cellular components of neuro-muscular junctions) during physiological and pathological perturbations of skeletal muscle homeostasis.
We use molecular, biochemical and epigenetic tools to understand structural and functional principles of the 3D genome organization that regulates gene expression during muscle regeneration and diseases
A topic of particular interest is the analysis of chromatin interactions that define the 3D genome organization and the identification of structural and functional interactions that regulate cell type-specific patterns of gene expression in response to cues released within the skeletal muscle regenerative environment in health and disease conditions, such as muscular dystrophies and other neuromuscular diseases.
The knowledge derived from our studies is instrumental to elucidate the pathogenesis of muscular disorders and discover pharmacological interventions that promote muscle regeneration to repair diseased muscles
Current translational focus is devoted to:
- the study of the therapeutic potential of HDAC inhibitors for treatment of Duchenne Muscular Dystrophy (DMD)
- the identification of genome variants associated to DMD patient-specific patterns of expression of disease-modifier genes that can account for individual trends of disease progression beyond the common genetic deficiency of dystrophin
- the effect of dystrophin deficiency and restoration by gene therapy on 3D genome and transcriptional output of DMD myofibers; the therapeutic potential of extracellular vesicles released by fibro-adipogenic progenitors of DMD skeletal muscles exposed to HDACi.
1. Epigenetic regulation of skeletal myogenesis by histone acetyltransferases and deacetylases
Our earlier identification and characterization of acetyltransferases p300/CBP and PCAF, as transcriptional co-activators, and the histone deacetylases HDACs, as transcriptional co-repressors, of the myogenic determination factor MyoD1-3, respectively, inspired the experimental rationale toward exploiting pharmacological inhibition of HDAC to promote skeletal myogenesis.
- p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. EMBO J 1997 Jan 15 ;16(2):369-8
- Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M. Mol Cell 1997 Dec ;1(1):35-45
- Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V. Mol Cell 2001 Oct ;8(4):885-97
- Stage-specific modulation of skeletal myogenesis by inhibitors of nuclear deacetylases. Iezzi S, Cossu G, Nervi C, Sartorelli V, Puri PL. Proc Natl Acad Sci U S A 2002 May 28 ;99(11):7757-62
- Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Iezzi S, Di Padova M, Serra C, Caretti G, Simone C, Maklan E, Minetti G, Zhao P, Hoffman EP, Puri PL, Sartorelli V. Dev Cell 2004 May ;6(5):673-84
2. HDAC inhibitors as pharmacological intervention in DMD and other muscular dystrophies
Puri lab discovered that dystrophin-activated nNOS signalling controls HDAC2 activity, thereby revealing a previously unrecognized link between constitutive activation of HDAC2 and alteration of the epigenetic landscape of dystrophin-deficient muscles6,7. This discovery established the rationale for using HDAC inhibitors to counter the progression of Duchenne muscular dystrophy (DMD), by correcting aberrant HDAC activity in dystrophin-deficient muscles8-11.
- Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Minetti G. C., Colussi c., Adami R., Serra C., Mozzetta C., Parente V., Illi B., Fortuni S., Straino S., Gallinari P., Steinkhuler C., Capogrossi M., Sartorelli V., Bottinelli R., Gaetano C., Puri P.L. Nat. Med 2006 Oct ;12(10):1147-50.
- A Common Epigenetic Mechanism Underlies Nitric Oxide Donors and Histone Deacetylase Inhibitors Effect in Duchenne Muscular Dystrophy. Colussi C., Mozzetta C., Gurtner A. , Illi B., Straino S., Ragone G., Pescatori M., Zaccagnini G., Rosati G., Minetti G., Martelli F., Ricci E., Piaggio G., Gallinari P., Steinkulher C., Capogrossi M.C., Puri P.L*, Carlo Gaetano. Proc Natl Acad Sci U S A 2008 Dec 9 ;105(49):19183-7 *Corresponding author.
- Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old mdx mice. Mozzetta C., Consalvi S., Saccone V., Tierney M., Diamantini A., Mitchel K.J., Marazzi G., Borsellino G., Battistini L., Sassoon D., Sacco A., Puri P.L. EMBO Mol Med 2013 Apr ;5(4):626-39
- Histological effects of givinostat in boys with Duchenne muscular dystrophy. Neuromuscul Disord. Bettica P, Petrini S, D’Oria V, D’Amico A, Catteruccia M, Pane M, Sivo S, Magri F, Brajkovic S, Messina S, Vita GL, Gatti B, Moggio M, Puri PL, Rocchetti M, De Nicolao G, Vita G, Comi GP, Bertini E, Mercuri E. Neuromuscul Disord 2016 Oct ;26(10):643-649
- HDAC inhibitors tune miRNAs in extracellular vesicles of dystrophic muscle-resident mesenchymal cells. Sandonà M, Consalvi S, Tucciarone L, De Bardi M, Scimeca M, Angelini DF, Buffa V, D’Amico A, Bertini ES, Cazzaniga S, Bettica P, Bouché M, Bongiovanni A, Puri PL, Saccone V. EMBO Rep 2020 Sep 3 ;21(9):e50863
- Determinants of epigenetic resistance to HDAC inhibitors in dystrophic fibro-adipogenic progenitors. Consalvi S, Tucciarone L, Macrì E, De Bardi M, Picozza M, Salvatori I, Renzini A, Valente S, Mai A, Moresi V, Puri PL. EMBO Rep 2022 Jun 7 ;23(6):e54721
3. Control of chromatin structure in muscle cells by regeneration-induced signaling pathways
Upon the discovery and characterization of intracellular signaling pathways (i.e. p38, ERK and AKT cascades) that regulate muscle gene expression in myoblasts, in earlier studies during Puri’s postdoctoral training, Puri lab has revealed the mechanism by which muscle environmental cues are converted into epigenetic changes that regulate gene expression in healthy and diseased muscles, via extracellular signal-activated kinase targeting of chromatin-modifying enzymes. These studies provided the first evidence that regeneration activated p38 and AKT signaling cooperatively direct assembly and activation of histone acetyltransferases and chromatin remodeling SWI/SNF complex at myogenic loci in muscle progenitors12,13,15. Moreover, we discovered that regeneration-activated p38 targets Polycomb Repressory Complex (PCR2) at Pax7 locus to promote formation of repressive chromatin during satellite cells a ctivation14.
- p38 Pathway Targets SWI/SNF Chromatin Remodeling Complex to Muscle-Specific Loci. Simone C., Forcales S.V., Hill D., Imbalzano A.L., Latella L., and Puri P.L. Nat Genet 2004 Jul ;36(7):738-43
- Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/Pi3K/AKT pathways during muscle differentiation. Serra C., Palacios D., Mozzetta C Forcales S., Ripani M., Morantte I., Jones D. Du K., Jahla U., Simone C., Puri P.L. Mol Cell 2007 Oct 26 ;28(2):200-13
- TNF/p38 alpha/Polycomb signalling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Palacios D., Mozzetta C., Consalvi S., Caretti G., Saccone V., Proserpio V., Marquez V.E., Valente S., Mai A., Forcales S., Sartorelli V., Puri P.L. Cell Stem Cell 2010 Oct 8 ;7(4):455-69
- Signal dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodeling complex. Forcales S., Albini S., Giordani L., Malecova B., Cignolo L., Chernov A., Coutinho P., Saccone V., Consalvi S., Williams R., Wang K., Wu Z., Baranovskaya S., Miller A., Dilworth F., Puri P.L. EMBO J 2012 Jan 18 ;31(2):301-16
4. Epigenetic basis for activation of the myogenic program in ESCs and other pluripotent cell types
Puri lab studied the epigenetic determinants of human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs) commitment to skeletal myogenesis, by investigating the hESC resistance to direct conversion into skeletal muscle upon ectopic expression of MyoD, which can otherwise reprogram somatic cells into the skeletal muscle lineage. These studies showed that hESC and hiPSC resistance to myogenic conversion is caused by the lack of expression of one structural component of the SWI/SNF chromatin remodelling complex – BAF60C – which is specifically induced in embryoid bodies13. Based on these studies, we have recently established a protocol of hESC-derived 3D contractile myospheres that offers the unprecedented opportunity to dissect and analyze the epigenetic dynamics that underlie the formation of skeletal muscles and to identify changes in the epigenome induced by contractile activity in healthy vs dystrophin-deficient myofibers16,20. We have also determined the identity of the general transcription factors implicated in the activation of skeletal myogenesis17, and we have discovered that replicative senescence is associated with acquisition of resistance to MYOD-mediated activation of muscle gene expression, caused by the constitutive activation of DNA damage repair (DDR) response that impairs cell cycle progression and MYOD activity18. Finally, our recent work has elucidated the mechanism by which MYOD regulates high-order chromatin interactions to define the tri-dimensional (3D) nuclear architecture for the activation of skeletal myogenesis during human somatic cell reprogramming into skeletal muscles19.
- Epigenetic reprogramming of human embryonic stem cells (hESCs) into skeletal muscle cells and generation of contractile myospheres. Albini S., Coutinho P., Malecova B., Giordani L., Savchenko A., Forcales S, Puri P.L. Cell Rep 2013 Mar 28;3(3):661-70
- TBP/TFIID-dependent activation of MyoD target genes in skeletal muscle cells. Malecova B., Dall’Agnese A., Madaro L., Gatto S., Coutinho Toto P., Albini S., Ryan T., Tora L., Puri PL. Elife 2016 Feb 25 ;5:e12534
- 18) DNA damage signaling mediates the functional antagonism between replicative senescence and terminal muscle differentiation. Latella L., Dall’Agnese A, Boscolo F., Nardoni C. Cosentino M., Lahm A, Sacco A., Puri P.L. Genes Dev 2017 Apr 1 ;31(7):648-659
- Transcription Factor-Directed Re-Wiring of Chromatin Architecture for Somatic Cell Nuclear Reprogramming Toward Trans-differentiation. Dall’Agnese A., Caputo L., Nicoletti C., di Iulio J., Schmitt A., Gatto S., Diao Y., Ye Z., Forcato M., Perera R., Bicciato S., Telenti A., Ren B., Puri P.L. Mol Cell 2019 Nov 7 ;76(3):453-472.e8
- Acute conversion of patient-derived Duchenne muscular dystrophy iPSC into myotubes reveals constitutive and inducible over-activation of TGFβ-dependent pro-fibrotic signaling. Caputo L, Granados A, Lenzi J, Rosa A, Ait-Si-Ali A, Puri PL, Albini S. Skelet Muscle 2020 May 2 ;10(1):13
5. Identification, functional, phenotypic and molecular characterization of muscle-interstitial cells – (the fibroadipogenic progenitors – FAPs) in healthy and diseased muscles.
Our work has elucidated the molecular determinants of the interplay between adult muscle stem cells and cellular components of their functional niche (i.e. FAPs), by identifying regulatory networks implicated in compensatory or pathogenic regeneration, and suggesting “disease stage-specific” responses to pharmacological treatment of neuromuscular disorders, such as DMD. Indeed, we have shown that HDACi promote compensatory regeneration and prevent fibro-adipogenic degeneration in mdx mice at early stages of diseases, by targeting a population of muscle interstitial cells – FAPs8 – and have identified a HDAC-regulated network that controls expression of myomiRs and alternative incorporation of BAF60 variants into SWI/SNF complexes to direct the pro-myogenic or fibro-adipogenic FAP activity21. Furthermore, we have recently identified specific subpopulations of FAPs (subFAPs) in physiological conditions and disease22 and we have discovered that specific subFAPs expand and adopt pathogenic phenotypes upon muscle denervation23 or in muscles of patients affected by type 2 diabetes24.
- HDAC-regulated myomiRs control BAF60 variant exchange and direct the functional phenotype of fibro-adipogenic progenitors in dystrophic muscles. Saccone V, Consalvi S, Giordani L, Mozzetta C, Barozzi I, Sandoná M, Ryan T, Rojas-Muñoz A, Madaro L, Fasanaro P, Borsellino G, De Bardi M, Frigè G, Termanini A, Sun X, Rossant J, Bruneau BG, Mercola M, Minucci S, Puri P.L. Genes Dev 2014 Apr 15 ;28(8):841-57
- Spectrum of cellular states within Fibro-Adipogenic Progenitors upon physiological and pathological perturbations of skeletal muscle. Malecova B., Gatto S., Etxaniz U, Passafaro M. Cortez A., Nicoletti C., Giordani L., Torcinaro A., De Bardi M., Bicciato S., De Santa F., Madaro L, Puri PL. Nat Commun 2018 Sep 10 ;9(1):3670
- Denervation-activated STAT3-IL6 signaling in fibro-adipogenic progenitors (FAPs) promotes myofibers atrophy and fibrosis. Madaro L, Passafaro M., Sala D., Etxaniz U., Lugarini F, Proietti D., Alfonsi MV, Nicoletti C., Gatto S., De Bardi M., Rojas-García R, Giordani L., Marinelli S., Pagliarini V., Sette C, Sacco A, Puri PL. Nat Cell Biol 2018 Aug ;20(8):917-927
- Human skeletal muscle CD90+ fibro-adipogenic progenitors are associated with muscle degeneration in type 2 diabetic patients. Farup J, Just J, de Paoli F, Lin L, Jensen JB, Billeskov T, Roman IS, Cömert C, Møller AB, Madaro L, Groppa E, Fred RG, Kampmann U, Gormsen LC, Pedersen SB, Bross P, Stevnsner T, Eldrup N, Pers TH, Rossi FMV, Puri PL, Jessen N. Cell Metab 2021 Nov 2 ;33(11):2201-2214.e11
 Mar 12, 2025
Mar 12, 2025Meet the La Jolla researcher who helped discover new muscular dystrophy drug
Mar 12, 2025People in Your Neighborhood: Dr. Pier Lorenzo Puri of Sanford Burnham Prebys says he was ‘obsessed with doing something’ to…
 Nov 13, 2024
Nov 13, 2024Decades of dedication led to FDA approval of a new treatment for Duchenne Muscular Dystrophy
Nov 13, 2024Nearly 30 years of discoveries by a Sanford Burnham Prebys scientist and collaborators lead to federal approval of the first…
 Dec 14, 2020
Dec 14, 2020Our top 10 discoveries of 2020
Dec 14, 2020This year required dedication, patience and perseverance as we all adjusted to a new normal—and we’re proud that our scientists
 Sep 14, 2020
Sep 14, 2020Scientists uncover a novel approach to treating Duchenne muscular dystrophy
Sep 14, 2020The study reveals a promising new therapeutic approach for the incurable muscle-wasting condition.
 Sep 10, 2019
Sep 10, 2019How your DNA takes shape makes a big difference in your health
Sep 10, 2019The more we learn about our genome, the more mysteries arise. For example, how can people with the same disease-causing…
Lukas Chavez is an Associate Professor at the Sanford Burnham Prebys. He is also the Director of the Clayes Research Center for Neuro-Oncology at the Institute for Genomic Medicine at the Rady Children’s Hospital, San Diego. In this role, he works with a team of physicians and scientists to capture genomic, transcriptomic, epigenetic and functional data from pediatric brain tumor patients, and uses this information to improve diagnosis and treatment. His research interests focus on structural variants as well as circular extrachromosomal DNA (ecDNA) in childhood cancers. These extrachromosomal DNA circles are frequently found in highly aggressive solid tumors and represent a new target for improved therapeutic approaches.
Education
2010: PhD, Free University, Berlin
Honors and Recognition
2020: St. Baldrick’s Scholar Award, St. Baldrick’s Foundation
2019: Award of Excellence in Pediatric Neuro-Oncology, Society of Neuro-Oncology
2012–2015: Feodor-Lynen Fellowship for Postdoctoral Researchers, Alexander-von-Humboldt Foundation
Related Disease
Brain Cancer, Cancer
Phenomena or Processes
Cancer Biology, Cancer Epigenetics, Chromosome Dynamics, Combinatorial Therapies, Gene Regulation, Genomic Instability, Oncogenes, Transcriptional Regulation
Anatomical Systems and Sites
Brain
Research Models
Clinical and Transitional Research, Computational Modeling, Cultured Cell Lines, Human, Human Cell Lines, Mouse
Techniques and Technologies
Bioinformatics, Cell Biology, Computational Biology, Computational Modeling, Gene Expression, Gene Silencing, Genomics, Single Nucleotide Polymorphisms (SNPs)
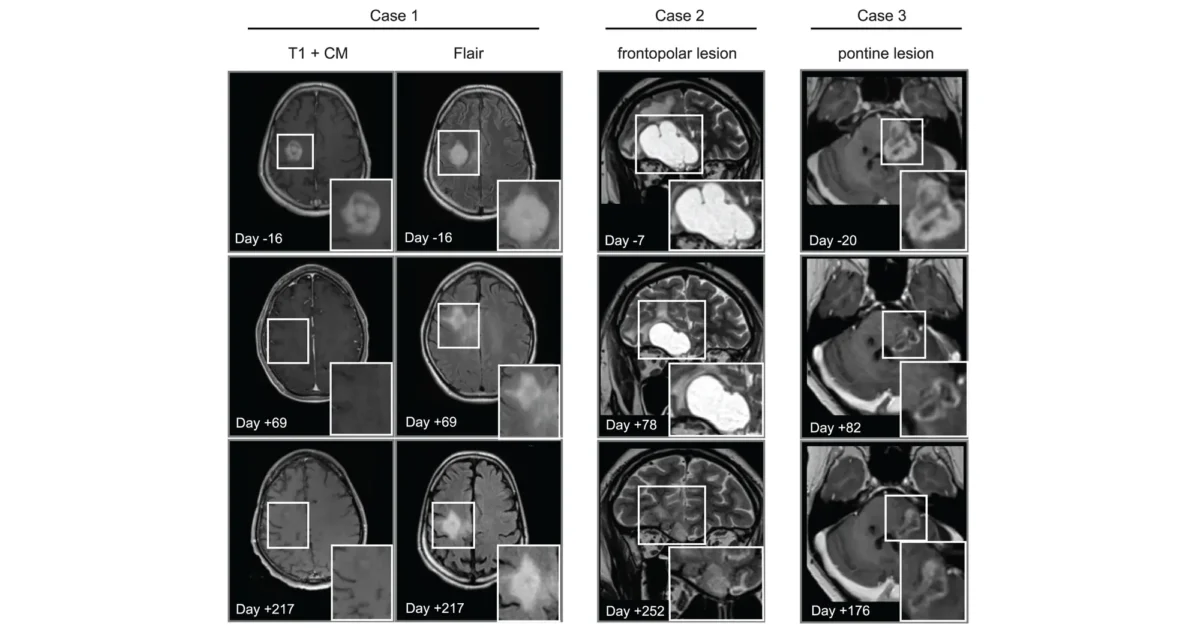 Apr 14, 2025
Apr 14, 2025Cancer drug finds new purpose in the brain
Apr 14, 2025Scientists show that a cancer drug travels to and shrinks some brain tumors, which may lead to new therapies.
 Aug 6, 2024
Aug 6, 2024Coding clinic
Aug 6, 2024Rapidly evolving computational tools may unlock vast archives of untapped clinical information—and help solve complex challenges confronting healthcare providers
 Jul 30, 2024
Jul 30, 2024Using machines to personalize patient care
Jul 30, 2024Artificial intelligence (AI) and other computational techniques are aiding scientists and physicians in their quest to create treatments for individuals…
 Jul 17, 2024
Jul 17, 2024Sanford Burnham Prebys announces new faculty recruit and two faculty promotions
Jul 17, 2024Douglas Sheffler was named as a new associate professor at Sanford Burnham Prebys, while Cosimo Commisso and Nicholas Cosford garnered…
 Mar 25, 2024
Mar 25, 2024Seminar Series: extrachromosomal DNA and the metabolic circuits of cancer immune suppression
Mar 25, 2024The ongoing Sanford Burnham Prebys seminar series will feature a pair of speakers on March 27, from noon to 1p.m.,…
 Nov 14, 2023
Nov 14, 2023“DNA loops” in pediatric brain tumors double relapse risk
Nov 14, 2023Findings help explain why some medulloblastoma tumors resist standard treatment.