Scientist Position: Member
Related Disease
HIV/AIDS, Infectious Diseases, Molecular Biology
Phenomena or Processes
Host-Pathogen Interactions, Infectious Disease Processes, Inflammation, Innate Immunity
Anatomical Systems and Sites
Immune System and Inflammation
Research Models
Clinical and Transitional Research, Computational Modeling, Human, Human Cell Lines, Mouse, Mouse Cell Lines, Primary Cells, Primary Human Cells
Techniques and Technologies
Biochemistry, Bioinformatics, Cellular and Molecular Imaging, Drug Discovery, Drug Efficacy, Gene Expression, Gene Knockout (Complete and Conditional), Gene Silencing, High-Throughput/Robotic Screening, RNA Interference (RNAi), Systems Biology
Education
University College Dublin
BSc and PhD, Pharmacology, cell and molecular biology, signal transduction and cancer, First Class Honors
Related Disease
Brain Cancer, Breast Cancer, Leukemia/Lymphoma, Lung Cancer, Pancreatic Cancer, Skin Cancer and Melanoma
Dr. Finlay’s research is focused on apoptosis (programmed cell death) and how this process can be manipulated to kill cancer cells.
Before joining Sanford Burnham Prebys, Tim was a scientist in Inflammation and Immunology Research at Celgene, San Diego. He received his PhD in Biochemistry and Molecular Biology from the Australian National University and completed postdoctoral training at La Jolla Institute for Allergy and Immunology.
Related Disease
Inflammatory/Autoimmune Disease
Tim works in the Ware lab where his research focus is the therapeutic potential of LIGHT-HVEM-BTLA co-signaling pathways in autoimmune-mediated inflammatory disorders and cancer immunotherapy.
Dr. Wechsler-Reya’s research focuses on the signals that control growth and differentiation in the cerebellum, and how these signals are dysregulated in the brain tumor medulloblastoma. As a postdoc, he demonstrated that Sonic hedgehog (Shh) is a critical mitogen for neuronal precursors in the cerebellum, and that mutations in the Shh pathway predispose to medulloblastoma by activating a mitogenic pathway that normally functions only in early development.
Now in his own lab, he continues to study the relationship between brain development and brain tumor formation. His lab’s contributions include identifying N-myc as a key target of the Shh pathway in neuronal precursors and in tumor cells; discovering a novel population of neural stem cells in the neonatal cerebellum; demonstrating that both neuronal precursors and stem cells can serve as cells of origin for MB; and identifying a population of cancer stem cells that is critical for propagation of Shh-associated tumors.
More recently, Dr. Wechsler-Reya and his group have begun developing new models of medulloblastoma and are using them to test novel therapeutic approaches. His work has garnered several awards, including a Kimmel Scholar Award, an Award for Excellence in Pediatrics Research from the Society for Neuro-Oncology and a Leadership Award from the California Institute for Regenerative Medicine (CIRM).
Education
2001-2010: Associate Professor of Pharmacology and Cancer Biology, Duke University Medical Center
1997-2001: Postdoctoral Fellow, Stanford University, Neural Development
1995-1996: Postdoctoral Fellow, Wistar Institute, Molecular Oncology
1995: PhD, University of Pennsylvania, Immunology
1986: B.A., Harvard College, Psychology & Biology
Funding Awards and Collaborative Grants
Leadership Award from the California Institute for Regenerative Medicine (CIRM)
Other Affiliations
2012: 19th International Brain Tumor Research and Therapy Conference, University of Toronto, Niagara Falls, ON
2012: “Developmental tumors of the nervous system,” held in Barcelona on July 2012, as part of the 8th Forum of European Neuroscience Societies.
Honors and Recognition
2007: W.K. Joklik Award for Excellence in Basic Cancer Research
2007: DukeMed Scholar
2006: Award for Excellence in Pediatrics Research, Society for Neuro-Oncology
2003: Kimmel Scholar Award, Sidney Kimmel Foundation for Cancer Research
2003: Brain Tumor Society Research Award
2002: Children’s Brain Tumor Foundation Research Award
2000-2001: Postdoctoral Fellowship, American Cancer Society (California)
1995-1997: Postdoctoral Fellowship, Medical Research Council of Canada
1988: Award for Excellence in Scientific Writing, American Diabetes Association
1984-1985: John Harvard Scholarship for Academic Achievement of Highest Distinction
Related Disease
Brain Cancer, Childhood Diseases
The Wechsler-Reya Lab studies the signals that control cell growth and differentiation in the nervous system and how these signals are dysregulated in brain tumors. We focus on medulloblastoma, the most common malignant brain tumor in children, and use models to understand the disease and to develop novel approaches to therapy. Our current areas of interest include:
- Discovering oncogenic drivers and creating new models
- Elucidating the molecular mechanisms of metastasis
- Identifying new therapeutics and approaches to drug delivery
- Harnessing the immune system to target tumors
We also work closely with physicians at Rady Children’s Hospital and elsewhere to translate our findings into trials that can benefit patients. Our goal is to develop safer and more effective therapies for children with brain tumors.
Robert Wechsler-Reya’s Research Report
Discovering Oncogenic Drivers and Creating Models
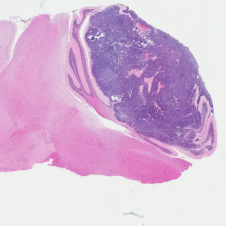
H&E stained Myc-Gfi1 tumor
Aggressive therapies – including surgery, radiation and high-dose chemotherapy – have improved outcomes for medulloblastoma patients, but many patients still die of their disease, and survivors suffer severe long-term side effects from therapy. To develop safer and more effective treatments, we need to understand the genes and pathways that are important for tumorigenesis. But for many forms of medulloblastoma, the oncogenic drivers are still unknown. A major focus of our research is to identify these drivers and use them to create robust animal models of the disease. Sequencing studies have identified genes that are altered in human medulloblastoma, and we are using functional assays to determine which of these genes can promote tumor growth in vivo. We have also established a large bank of patient-derived xenograft models that we use to perturb candidate genes and test their roles in tumorigenesis. In addition to identifying new drivers of medulloblastoma, these studies generate models that can be used to test novel approaches to therapy.
Elucidating the Molecular Mechanisms of Metastasis
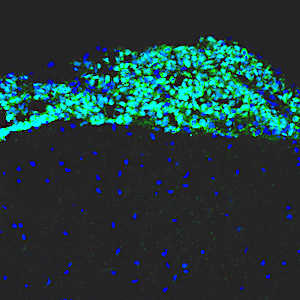
Leptomenigeal metastasis section
Most medulloblastoma research has focused on the primary tumors growing in the cerebellum. However, most medulloblastoma patients do not die from the primary disease, but from leptomeningeal metastasis: the dissemination of tumor cells from the cerebellum into the brain and spinal cord. Metastatic lesions cannot be surgically removed, and there are no effective therapies to eliminate them or stop them from growing. To understand the molecular basis of metastasis, we are using animal models to study the differences between primary and metastatic tumors, and screening for genes that can promote or inhibit metastatic dissemination. We are also integrating our results with molecular data from medulloblastoma patients, to identify genes that are critical for metastasis. Understanding the function of these genes will allow us to design novel strategies for early detection, prevention, and treatment of metastasis in patients with medulloblastoma and other types of brain tumors.
Identifying new Therapeutics and Approaches to Drug Delivery

Tumor-homing peptides in MP tumor
Genomic analyses have revealed that medulloblastoma is an extremely heterogeneous disease, with at least 4 distinct subtypes that differ in terms of mutations, gene expression, epigenetic changes, and patient survival. Despite this heterogeneity, most medulloblastoma patients currently receive the same therapy. A major goal of our research is to discover new therapeutic strategies that are tailored to specific medulloblastoma subgroups. To this end, we have assembled a large panel of patient-derived xenografts and are using them for high-throughput drug screening. Working with experts in genomics and computational biology, we are using statistical and mathematical tools to understand the relationship between molecular alterations and drug responses. These studies not only highlight new targeted therapies for medulloblastoma, but also provide insight into drug response biomarkers and help prioritize agents for clinical trials.
In addition to identifying therapeutics, we are also exploring novel approaches to drug delivery. A major obstacle to treating brain tumors is that the majority of small molecule drugs are not able to cross the blood brain barrier, and those that do are often pumped out by multi-drug transporters. To solve this problem, we are collaborating with bioengineers with expertise in nanotechnology. By encapsulating drugs in nanoparticles and delivering them directly to the central nervous system, we hope to increase the concentration of drugs in brain tumors and reduce the concentrations in other tissues, thereby mitigating systemic side effects. We have already identified several drugs that are effective at killing medulloblastoma cells in vitro; if we can develop strategies for effective delivery of these drugs to tumors, we can markedly improve outcomes for medulloblastoma patients.
Harnessing the Immune System to Target Tumors
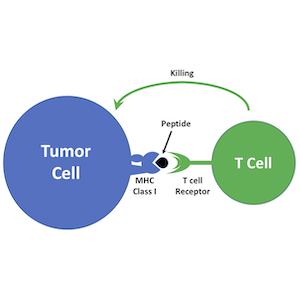
Immunotherapy is emerging as a powerful approach to treating cancer. Antagonists of immune checkpoint regulators, T lymphocytes engineered to recognize tumor antigens, and vaccines that amplify tumor-specific lymphocytes are being tested against a variety of human malignancies. Although some remarkable successes have been reported, only a subset of patients respond to these therapies, and the mechanisms that underlie resistance are poorly understood. Pediatric brain tumors, in particular, have not yet benefited from immunological targeting. We are studying the mechanisms brain tumors use to evade the immune system and suppress immune responses, and developing therapeutic strategies to overcome these mechanisms. We are also using genomic and proteomic approaches to identify antigens that might represent novel targets for vaccines, CAR T cells and natural killer cells. Finally, we are “humanizing” our PDX models so we can explore interactions between the immune system and patient-derived tumor cells. By increasing the immunogenicity of tumor cells and enhancing anti-tumor immune responses, we hope to bring the benefits of immunotherapy to medulloblastoma patients.
 Jul 6, 2022
Jul 6, 2022Heating up cold brain tumors: An emerging approach to medulloblastoma
Jul 6, 2022Immunotherapy has revolutionized cancer treatment, but it doesn’t work on many childhood brain tumors. Researchers from Sanford Burnham Prebys are…
 Apr 7, 2021
Apr 7, 2021Conrad Prebys Foundation provides $3 million for pediatric brain cancer research
Apr 7, 2021Conrad Prebys was an extraordinary man and a passionate philanthropist. Today, his generosity extends beyond his life through the Conrad…
 Dec 14, 2020
Dec 14, 2020Our top 10 discoveries of 2020
Dec 14, 2020This year required dedication, patience and perseverance as we all adjusted to a new normal—and we’re proud that our scientists
 Nov 9, 2020
Nov 9, 2020Personalized drug screens could guide treatment for children with brain cancer
Nov 9, 2020A clinical trial that evaluates the approach for medulloblastoma, the most common malignant pediatric brain tumor, is planned Scientists at…
 Jul 9, 2020
Jul 9, 2020Sanford Burnham Prebys researchers awarded 2020 Padres Pedal the Cause grants
Jul 9, 2020We are pleased to announce that Padres Pedal the Cause (PPTC) has awarded three collaborative research grants to Sanford Burnham…
 Jan 13, 2020
Jan 13, 2020How to help children survive—and thrive—after a brain cancer diagnosis
Jan 13, 2020Lynne Selinka knew in her heart that something was seriously wrong with her 10-year-old son, Travis. For months he had…
Dr. Hauser earned his PhD in Molecular Genetics from University of California at Irvine. Following postdoctoral training at UC Berkeley, Dr. Hauser was recruited to the Sanford-Burnham faculty in 1989, and was promoted to Associate Professor in 1996. In 2005, he became an adjunct faculty member and assumed a full-time administrative role. Dr. Hauser currently serves as Vice President for Scientific Resources, and as Cancer Center Associate Director, Shared Resources, overseeing Sanford Burnham Prebys’ Shared Resource operations, scientific equipment, and scientific regulatory compliance.
Related Disease
Cancer
Dr. Hauser’s research has centered on the interplay between the regulation of gene expression and oncogenic transformation. A major focus has been determining the role of the Ets family of transcription factors in cancer. His work has defined several distinct roles for Ets factors in mediating the many features of cellular transformation, and revealed unanticipated complexity in this transcription factor gene family.
Dr. Zhao joins us from University of California San Francisco, where he recently completed his Postdoctoral Training. His lab will focus on understanding how proteins function under different physiological and disease states from a structural biology perspective. Specifically, Dr. Zhao brings significant expertise in visualizing proteins at high resolution using cryogenic electron microscopy (cryo-EM).
Dr. Zhao received his Bachelor’s and PhD in Medical Biophysics from the University of Toronto, Canada, where he completed 5 years of graduate training investigating rotary ATPases. He then went on and completed 5 years of postdoctoral training at UCSF studying Transient Receptor Potential ion channels.
How to build our body’s protein recycling factories
Sanford Burnham Prebys scientists have learned more about the cell’s recycling process for obsolete and misshapen proteins.
Dr. Yip is professor and director of the Center for Data Science and Artificial Intelligence at Sanford Burnham Prebys Medical Discovery Institute. A leader in computational biology and bioinformatics at Chinese University of Hong Kong, he was recruited in 2022 to further elevate and accelerate Sanford Burnham Prebys’ growing capabilities and ambitions in next-generation biomedical research tools and approaches.
For almost 20 years, Dr. Yip’s research has focused on three primary interests: development of computational methods for analyzing data produced by emerging experimental technologies, such as single-cell and spatial transcriptomics; studying fundamental gene regulatory mechanisms using machine learning and data science methods; and identifying, annotating and interpreting genomic, transcriptomic and epigenomic changes in human diseases, such as cancers, diabetes, and neurodegenerative diseases.
Under his leadership, the mission of the Center for Data Science and Artificial Intelligence is to effectively tap the almost unlimited potential of rapidly evolving large-scale data sets and computational tools in biomedical research, with an emphasis on interdisciplinary collaborations that leverage the expertise of many disciplines to reveal new actionable knowledge.
Education and Training
2010: Postdoctoral associate, Molecular Biophysics and Biochemistry, Yale University
2009: PhD, Computer Science, Yale University
2003: M.Phil., Computer Science, The University of Hong Kong
1999: B.Eng., Computer Engineering, The University of Hong Kong
Related Disease
Biliary Atresia, Cancer, Diabetes – General, Hirschsprung Disease, Liver Cancer, Nasopharyngeal Carcinoma, Type 2 Diabetes
Phenomena or Processes
Cancer Epigenetics, Gene Regulation, Oncogenes, Posttranslational Modification, Transcriptional Regulation, Tumor Microenvironment
Anatomical Systems and Sites
Endocrine System, General Cell Biology, Immune System and Inflammation, Liver
Research Models
Computational Modeling
Techniques and Technologies
Bioinformatics, Comparative Genomics, Genomics, Machine Learning, Protein-Protein Interactions, Systems Biology
The Yip lab studies gene regulatory mechanisms by means of computational modeling. To facilitate their data-centric approach, they develop novel methods for analyzing large amounts of biological data, including those produced by cutting-edge high-throughput experiments. Their computational models provide a systematic way to investigate the functional effects of different types of perturbations to regulatory mechanisms, which creates testable hypotheses for studying human diseases and facilitates translational research.
 Jul 11, 2025
Jul 11, 2025Cutting to the core of how 3D structure shapes gene activity
Jul 11, 2025New method can measure how secluded genomic regions are in 3D space and then link 3D position to gene activity.
 Aug 27, 2024
Aug 27, 2024Simulating science or science fiction?
Aug 27, 2024In the Conrad Prebys Center for Chemical Genomics, simulation-based techniques help scientists find new potential treatments.
 Aug 13, 2024
Aug 13, 2024Dodging AI and other computational biology dangers
Aug 13, 2024Sanford Burnham Prebys scientists say that understanding the potential pitfalls of using artificial intelligence and computational biology techniques in biomedical…
 Aug 8, 2024
Aug 8, 2024Scripting their own futures
Aug 8, 2024At Sanford Burnham Prebys Graduate School of Biomedical Sciences, students embrace computational methods to enhance their research careers
 May 11, 2023
May 11, 2023New algorithm can predict diabetic kidney disease
May 11, 2023Researchers from Sanford Burnham Prebys and the Chinese University of Hong Kong have developed a computational approach to predict whether…
 Feb 9, 2022
Feb 9, 2022Bioinformaticist Kevin Yip joins Sanford Burnham Prebys
Feb 9, 2022Bioinformaticist Kevin Yip, PhD, has joined Sanford Burnham Prebys as a professor, where he will collaborate with other faculty across the
Yu Yamaguchi earned his MD from Tohoku University in Japan in 1981, followed by a PhD in 1985, and training in obstetrics and gynecology at the same institute. Dr. Yamaguchi came to Sanford Burnham Prebys for his postdoctoral training. He was appointed to the staff in 1991.
Honors and Recognition
The Humanitarian Scientific Achievement Award, The MHE Research Foundation
The Kushima Prize, The Alumni Association, Tohoku University School of Medicine
Other Affiliations
Member, Scientific and Medical Advisory Board, The MHE Research Foundation
Related Disease
Alzheimer’s Disease, Arthritis, Autism Spectrum Disorders, Bone Mineralization Disorders, Epilepsy, Multiple Hereditary Exostoses
The goal of research in the Yamaguchi laboratory is to understand the role of proteoglycans and glycosaminoglycans in the context of development and human disorders. The general strategy is to define the role of proteoglycans and glycosaminoglycans by characterizing the phenotype of mutant mice lacking the synthesis of individual glycosaminoglycans. Specifically, mutant mice lacking the Ext1 and Has genes have been created to study heparan sulfate and hyaluronan, respectively. Recent progress in genetic studies in humans and mice has begun to reveal that deficiencies in glycosaminoglycans can be the causes and/or confounding factors of human childhood disorders. The Yamaguchi lab is now working to clarify the molecular mechanisms of two such disorders (multiple hereditary exostoses and autism) in order to develop new medical treatments.
For more information on the impact of Dr. Yamaguchi’s work, read letters from the patients with MHE.
Yu Yamaguchi’s Research Report
What Are Proteoglycans and Glycosaminoglycans?
Proteoglycans are a family of glycoproteins consisting of a core protein and a various number of long sugar chains called glycosaminoglycans attached to the core protein (Fig. 1). There are four classes of glycosaminoglycans; heparan sulfate, chondroitin sulfate, keratan sulfate, and hyaluronan (hyaluronic acid). Heparin, the anticoagulant widely used in clinics, is a specialized form of heparan sulfate. Although there is ample circumstantial evidence that these glycosaminoglycans have important biological functions, a complete understanding of their function and their relevance to human diseases requires genetic animal models.
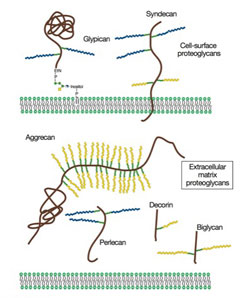
Figure 1. Proteoglycans consist of a protein core and one or more covalently attached glycosaminoglycan chains. From Esko, JD, Kimata, K., and Lindahl, U. Proteoglycans and Sulfated Glycosaminoglycans, In: Essentials of Glycobiology, CSH Press.
Developmental Roles Of Glycosaminoglycans
HEPARAN SULFATE – The Ext1 gene encodes an enzyme essential for the elongation of nascent heparan sulfate chains. As a result of genetic ablation of Ext1 using a ‘conditional knockout’ approach, heparan sulfate is eliminated from specific tissues and cell types. Our previous studies using brain-specific Ext1 conditional knockout have demonstrated critical roles of heparan sulfate in brain patterning, neurogenesis in the cerebral cortex, and pathfinding of various axon tracts (1)(2)(3)(4). More recently, conditional Ext1 knockout in developing limb bones revealed critical roles of heparan sulfate in the growth and patterning of bones and joint formation (5).
Moreover, our conditional Ext1 mutant mouse model has been distributed to more than 20 laboratories worldwide to characterize the role of heparan sulfate in various tissues and cell types, such as colon (6), kidney (7), lymphocytes (8), blood vessels (9), eyes (10), embryonic stem cells (11), and so on.
HYALURONAN – Three Has genes (Has1, Has2, Has3) encode the entire repertoire of hyaluronan synthases in mammalian cells. Genetic ablation of these genes, singly or in combination, results in a reduction or total elimination of hyaluronan, depending on the repertoire of Has expression in the given tissue. We created a conditional null allele of the Has2 gene, which is the predominant Has in many tissues. Our conditional Has2 knockout study targeted to the limb bud mesenchyme has revealed that hyaluronan plays a critical role in the proliferation and maturation of chondrocytes in the developing limb skeleton (12). Like Ext1 mutant mice, these Has2 conditional knockout mice are being used in more than a dozen laboratories worldwide for studies on the role of hyaluronan in various tissue and cell types.
Deficiencies in glycosaminoglycan synthesis can be the causes of childhood disorders
Recent progress in genetic studies in humans and mice has begun to reveal that deficiencies in glycosaminoglycans can be the causes and/or confounding factors of human childhood disorders. The research focus of our lab is to elucidate the molecular mechanisms of such disorders and to develop new medical treatments. We are currently studying two such disorders.
MULTIPLE HEREDITARY EXOSTOSES – One of the major diseases studied in our lab is Multiple Hereditary Exostoses (MHE; also known as Hereditary Multiple Exostoses [HME] or Multiple Osteochondroma [MO]). MHE is caused by a mutation in Ext1 (see above) or its related gene, Ext2. As mentioned above, these genes encode an enzyme necessary to produce heparan sulfate. MHE occurs in children of 0-12 years old. Although no comprehensive survey has been conducted, it is estimated that there are several thousand individuals affected by MHE in the US, which makes MHE one of the more prevalent among ’rare diseases’. Dr. Yamaguchi is a member of the scientific advisory board of the MHE Research Foundation and has been working to promote collaborations between basic scientists, academic physicians, and patient advocates.
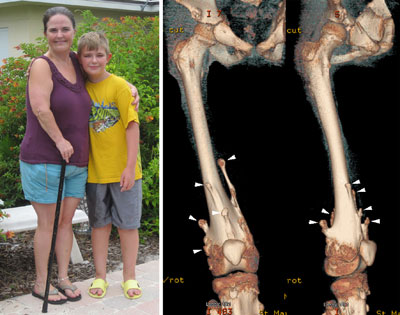
Figure 2. MHE patients, Carol and her 12-year-old son, Bruce. Shown on the right are three-dimensional CT images of Bruce’s right upper leg and knee joint area. Note that there are many bony protrusions (‘exostoses’), as indicated by white arrowheads. These tumors need to be surgically removed to prevent possible malignant transformation. Surgery is also needed to correct bone deformities and bone length inequalities. For example, Bruce and Carol have had 21 and 36 surgeries, respectively.
Children with MHE suffer from the formation of multiple –– sometimes as many as 100 –– bony tumors (osteochondromas) (Fig. 2). These bony tumors stunt their growth and can cause pain and disfigurement. Fortunately, the chance these tumors becomes cancerous is relatively low, partly because they are surgically removed as they develop. This means, however, children with MHE need to go through multiple surgeries over the course of their lives. There is currently no medical treatment for the disease.
Our lab is currently working to elucidate the molecular and cellular mechanisms of MHE. One of the major thrusts has been to create a mouse model that mimics the manifestations of human MHE. A long-term issue of MHE research has been the lack of mouse models that faithfully recapitulate the manifestation of human MHE; when Ext genes were inactivated in mice just as they are in human MHE patients, the mice failed to develop the symptoms of MHE.
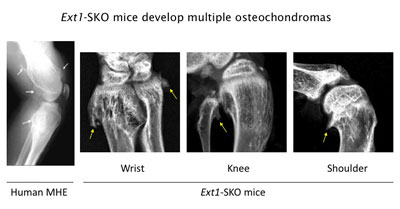
Figure 3. Ext1-SKO mice develop multiple osteochodromas in a pattern almost identical to human MHE. The X-ray images of the knee joint area of an MHE patient and the wrist, knee, and shoulder areas of Ext1-SKO mice. Osteochondromas are indicated by arrows. Ext1-SKO mice also mimic other skeletal deformities frequently seen human MHE, such as bowing of the forearm, the subluxation/dislocation of the radial head, and scoliosis (not shown).
Instead of knocking out the Ext1 gene in the whole mouse, we targeted the gene in only a small fraction of bone cells (Ext1-SKO mice). This minimalistic approach led to a mouse with all the physical manifestations of MHE, such as bony protrusions, short stature, and other skeletal deformities (Fig. 3)(13). The new mouse model answered some long-standing questions about MHE. Scientists had gone back and forth on whether osteochondromas observed in MHE are true tumors or just malformations of the bone. In this study, the tumors were made up of two cell types. A minority were mutant cells lacking Ext1, but, amazingly, most were normal bone cells. Hence, osteochondroma in MHE is not considered a true neoplasm in its strictest sense.
Our lab has been using this and additional mouse models to further dissect the pathogenic mechanism of MHE. Moreover, this mouse model provides new opportunities to test potential drugs to prevent osteochondroma formation and other clinical symptoms of MHE.
AUTISM – Children with MHE sometimes suffer from neurological and mental symptoms, which is not surprising because heparan sulfate is expressed and plays critical roles in the nervous system (1). The MHE community has long noticed the prevalence of autism-like behavioral traits in the patient population, and there are clinical reports describing the association of autism with MHE. Aided by the funds from the Sanford Health and the MHE Research Foundation, we have been studying the behavior of Ext1 mutant mice. Our preliminary data suggest that heparan sulfate has indeed a physiological function in the nervous system, and that its deficiency can cause behavioral deficits relevant to human autism. We are also analyzing DNA samples from individuals with autism for abnormalities in enzymes for heparan sulfate synthesis. Since heparan sulfate is a modulator of a number of neuronal molecules, we hope to identify functional networks of molecules underlying autism and other childhood mental disorders.
 Feb 1, 2022
Feb 1, 2022Sanford Burnham Prebys professor awarded $2.9 million to explore new answers to old questions in Alzheimer’s research
Feb 1, 2022Sanford Burnham Prebys professor Yu Yamaguchi, MD, PhD, has been awarded a $2.9 million grant from the National Institutes of Health
 Oct 21, 2021
Oct 21, 2021This enzyme is one of the hardest working proteins in the body
Oct 21, 2021Researchers have shown that a protein they identified plays a major role in the breakdown of hyaluronic acid, a compound…
 Mar 25, 2019
Mar 25, 2019Families find hope at our 10th Annual Rare Disease Day Symposium
Mar 25, 2019The unofficial theme of Sanford Burnham Prebys’ 10th annual Rare Disease Day symposium can be summarized in one word: hope.
 Jan 9, 2018
Jan 9, 2018Year in review: Top stories in 2017
Jan 9, 2018In the last 12 months, SBP scientists published 338 scientific papers—that’s almost a paper a day. We are proud of…
 Jul 14, 2017
Jul 14, 2017Genes and proteins go hand-in-hand
Jul 14, 2017Thanks to huge improvements in DNA sequencing technology, scientists have identified almost all the genes present in humans. Despite this…
 Mar 15, 2017
Mar 15, 2017Research may explain congenital deafness
Mar 15, 2017If you’ve heard of hyaluronic acid (HA), it’s probably as an ingredient in cosmetic products meant to help keep skin
Dr. Yu Xin (Will) Wang received his PhD at the University of Ottawa where he identified cellular asymmetry and polarity mechanisms regulating muscle stem cell self-renewal and skeletal muscle regeneration. He then carried out postdoctoral training at Stanford University School of Medicine developing single cell multi-omic approaches to characterize the regenerative process and what goes awry with disease and aging.
“I’ve always had a passion for science and became fascinated with how the body repairs and heals itself when I was introduced to the potential of stem cells in regenerative medicine. I was struck by the ability of a small pool of muscle stem cells that can rebuild and restore the function of muscle. My lab at Sanford Burnham Prebys aims to better understanding the repair process and harness our body’s ability to heal in order to combat chronic diseases and even counteract aging.“
Education and Training
Postdoctoral Fellowship, Stanford University School of Medicine
PhD in Cellular Molecular Medicine, University of Ottawa, Canada
BS in Biomedical Sciences, University of Ottawa, Canada
Prestigious Funding Awards
2020: NINDS K99/R00 Pathway to Independence Award
Honors and Recognition
Governor General’s Gold Medal – Canada
Related Disease
Aging-Related Diseases, Amyotrophic Lateral Sclerosis (Lou Gehrig’s Disease), Arthritis, Cachexia, Inflammatory/Autoimmune Disease, Multiple Sclerosis, Muscular Dystrophy, Myopathy, Neurodegenerative and Neuromuscular Diseases, Sarcopenia/Aging-Related Muscle Atrophy, Spinal Muscular Atrophy
Phenomena or Processes
Adult/Multipotent Stem Cells, Aging, Cell Signaling, Development and Differentiation, Epigenetics, Exercise, Extracellular Matrix, Neurogenesis, Organogenesis, Regenerative Biology, Transcriptional Regulation
Anatomical Systems and Sites
Immune System and Inflammation, Musculoskeletal System, Nervous System
Research Models
Clinical and Transitional Research, Computational Modeling, Human Adult/Somatic Stem Cells, Mouse
Techniques and Technologies
3D Image Analysis, Bioinformatics, Cellular and Molecular Imaging, Gene Knockout (Complete and Conditional), Genomics, High Content Imaging, High-Throughput/Robotic Screening, Live Cell Imaging, Machine Learning, Microscopy and Imaging, Proteomics, Transplantation
The Wang lab is interested in elucidating critical cell-cell interactions that mediate the function of tissue-specific stem cells during regeneration and disease, with a focus on how a coordinated immune response can promote regeneration and how autoimmunity impacts tissue function and hinder repair.
Specifically, the Wang lab aims to identify cellular and molecular crosstalk between muscle, nerve, and immune systems to develop targeted therapies that overcome autoimmune neuromuscular disorders and autoimmune aspects of “inflammaging.”
Yu Xin (Will) Wang’s Research Report
The lab’s research is translationally oriented and utilizes interdisciplinary molecular, genetic, computational (machine learning and neural networks), and bioengineering approaches to view biology and disease from new perspectives. We combine multi-omics sequencing and imaging methods to resolve how different cell types work together after injury to repair tissues and restore function. We use a data-driven approach to identify targetable disease mechanisms and, through collaborations with other researchers and clinicians, develop therapies that promote regeneration. Visit our lab website to learn more.
 Jun 12, 2025
Jun 12, 2025Turning back time on muscle stem cells to prevent frailty from aging
Jun 12, 2025Study from Dr. Will Wang’s lab finds a way to restore stem cells from aged muscle to become young again…
 Aug 20, 2024
Aug 20, 2024Mapping the human body to better treat disease
Aug 20, 2024Scientists are investigating the inner workings of our bodies and the cells within them at an unprecedented level of detail.
 Aug 13, 2024
Aug 13, 2024Dodging AI and other computational biology dangers
Aug 13, 2024Sanford Burnham Prebys scientists say that understanding the potential pitfalls of using artificial intelligence and computational biology techniques in biomedical…
 Oct 11, 2023
Oct 11, 2023Inhibiting an enzyme associated with aging could help damaged nerves regrow and restore strength
Oct 11, 2023New research has demonstrated a way to accelerate recovery from peripheral nerve injury by targeting an enzyme that was thought…
 Jan 26, 2023
Jan 26, 2023Three big questions for cutting-edge biologist Will Wang
Jan 26, 2023Will Wang’s spatial omics approach to studying neuromuscular diseases is unique.
 Nov 23, 2022
Nov 23, 2022Yu Xin (Will) Wang joins Sanford Burnham Prebys to advance regenerative medicine
Nov 23, 2022Molecular biologist Yu Xin (Will) Wang, PhD, has joined Sanford Burnham Prebys as an assistant professor in the Development, Aging,…