Overview
The Exploratory Pharmacology group delivers rapid, high-quality assessments of drug-like properties to support informed decision-making by medicinal chemistry teams during drug development. Leveraging deep scientific expertise and optimized workflow infrastructure, the group enhances efficiency across the drug discovery pipeline, from hit identification to candidate selection.
| Tier 1 ADME In vitro | Tier 2 ADME In vitro | Drug-like Properties In vivo |
|---|---|---|
| Kinetic Solubility | Cyp P450 inhibition | Analytical method development |
| Thermodynamic solubility | In vitro hERG binding | IV/IP/PO Pharmacokinetics |
| Microsomal Stability | Metabolite profiling | Oral bioavailability |
| Hepatocyte Stability | Plasma Protein Binding | Formulation screening |
| Plasma Stability | Tissue free fraction | Tissue/CNS Distribution |
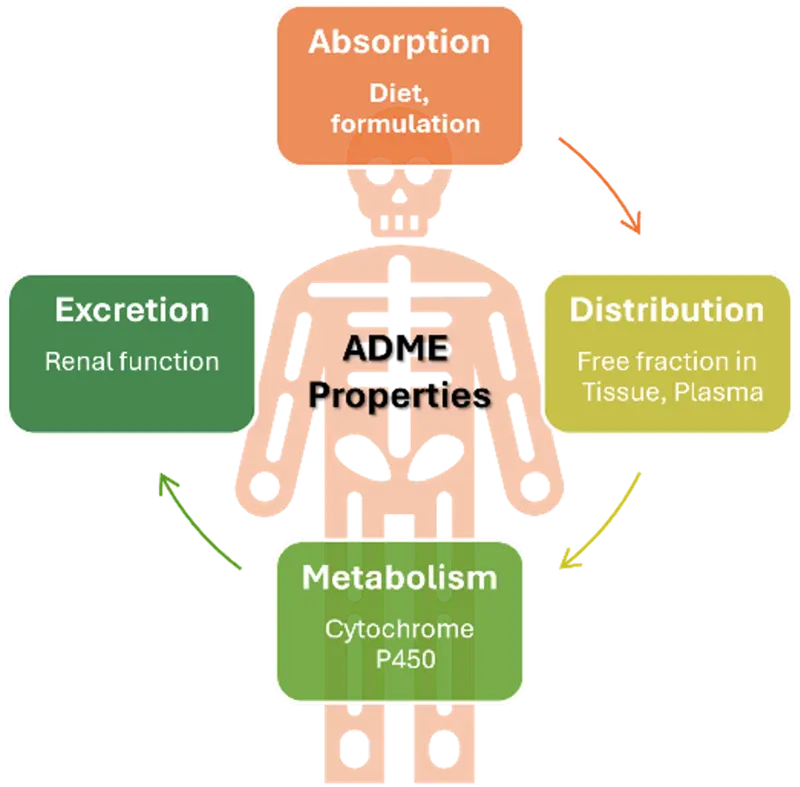
Comprehensive ADME Profiling
The bioanalytical facility performs critical drug discovery ADME (Absorption, Distribution, Metabolism, Excretion) assays, including:
- Solubility assessments under physiologically relevant conditions
- Metabolic stability studies in liver microsomes and hepatocytes (mouse, rat, human, minipig, dog and others on demand)
- Plasma protein and tissue binding experiments
- Cytochrome P450 inhibition screening
- Metabolite profiling and identification
Advanced Bioanalytical Capabilities
The team utilizes state-of-the-art Triple Quadrupole mass spectrometry (API 6500+ QTRAP) for ultra-sensitive quantification of drug concentrations in plasma/tissue samples. This technology enables analysis of exceedingly small sample volumes (5 µL), surpassing traditional PK protocols.
Highly efficient Micro-PK Study
A combination of saphenous, retro-orbital, and terminal bleeding techniques allows serial blood sampling (~10–15 µL per draw) from individual mice at up to 8 time points (0–24 hours post-dose). This micro-sampling approach, paired with high-sensitivity bioanalysis, generates complete PK profiles using only 3 mice per arm, minimizing animal use and reducing costs. Routinely conducted routes of administration include Intravenous (IV), Intraperitoneal (IP), Oral (PO), Subcutaneous (SC) and Topical administration routes.
The micro-PK procedure described is highly cost-effective as it minimizes the number of animals required while generating complete pharmacokinetic (PK) profiles from each subject. By employing micro-sampling techniques, only 3 mice per PK arm are used, significantly reducing animal use compared to traditional protocols. This approach not only lowers costs but also ensures ethical compliance by minimizing the number of animals needed for research. Additionally, the procedure utilizes advanced bioanalytical infrastructure, such as Triple Quad mass spectrometry, to extract comprehensive PK profiles from exceedingly small sample volumes, maximizing efficiency and accuracy.
External Study Support
The facility also analyzes samples from in vivo PK/PD studies performed outside of Prebys Center, offering:
- Drug level quantification via LC-MS/MS (Shimadzu UHPLC with Nexera SIL-30ACmp autosampler coupled to AB SCIEX API 6500+ QTRAP®/API 4000 systems) with a typical limits of quantification typically less than 1nM.
- Metabolite profiling
Extended Pharmacological Services
Additional capabilities include:
- Formulation screening for optimal bioavailability
- Allometric scaling and human dose predictions
- AI/ML-driven predictions for key pharmacological properties (e.g., brain penetration, CYP lability, P-gp interactions, metabolic pathways) for academic collaborators.
- We also have access to reliable vendors who perform high quality studies including permeability, CYP induction, met-ID, genotoxicity and other assays.
By integrating cutting-edge technology with specialized expertise, the Exploratory Pharmacology group accelerates the identification of viable drug candidates while adhering to ethical and cost-effective practices.
Services
Assays
- Stability
Liver microsome, hepatocyte, plasma and formulation stability - Solubility
Kinetic and thermodynamic solubility - Free fraction determination
Plasma protein binding and brain fractionation - PK and bioavailability studies
IV, IP, PO, SC, topical etc. - Dose predictions
Clearance, Brain weight and Maximum Lifespan Potential based First-in-Human predictions - PD and efficacy studies
In vivo models for assessing target engagement, functional activity and assessment of drug like properties
Equipment & Resources
Sciex QTRAP 6500+
Brings about significant enhancement in our bioanalysis capabilities
- Linearity: Triple quadrupole has one of the best range of linearity with MRM3 mode (up to 6 orders of magnitude)
- Sensitivity: LLOQ in pg/mL to fg/ml ranges (results vary for analytes; Attograms sensitivity reported for some analytes)
- Agility: Polarity switching (1millisecond)
- Versatility: Mass range of m/z 5 – 2,000
- Robustness: Multi-day consecutive injection feasible
- Efficiency: Increased ionization efficiency and heat transfer with the new IonDrive Turbo V source


Sciex API 4000
For ADME and bioanalysis
- Linearity: Triple quadrupole has one of the best range of linearity with MRM3 mode (up to 6 orders of magnitude)
- Sensitivity: LLOQ in ng/mL ranges (results vary for analytes; picograms sensitivity reported for some analytes)
- Agility: Polarity switching
- Versatility: Mass range of m/z 5 – 1,000
- Robustness: Multi-day consecutive injection feasible
- Efficiency: Increased ionization efficiency and heat transfer with the new IonDrive Turbo V source
Shimadzu UPLCs
Shimadzu UPLCs with advanced SIL-30 autosamplers are designed to deliver exceptional performance in bioanalytical experiments. These systems feature ultra-low carryover, rapid injection cycles, and precise sample handling, ensuring reliable and reproducible results. The needle-in-flow path design minimizes contamination by continuously flushing the sample path with the mobile phase gradient. With capabilities such as temperature-controlled sample cooling for heat-sensitive compounds, automated pretreatment functions, and dual injection modes, these autosamplers enhance throughput and precision while reducing manual intervention. Their compatibility with LC-MS/MS systems makes them ideal for high-sensitivity applications, enabling accurate quantification and profiling of analytes in complex biological matrices.
Price List
For a Price List, please email us.
Leadership
Raghuveer Ramachandra, PhD
Associate Director