Giovanni Paternostro earned his PhD in Biochemistry from the University of Oxford, England, in 1997. He has obtained his MD and Board Certification in Cardiology from the University of Rome, Italy. After postdoctoral training at the Imperial College School of Medicine, Hammersmith Hospital, London and at Sanford Burnham Prebys he was promoted to Research Investigator in 2001 and to Assistant Professor in 2003. In 2001 he was nominated member of the Whitaker Institute for Biomedical Engineering, UC San Diego. His research has been recognized by the 2002 Society for Geriatric Cardiology Basic Science Award and by the Ellison Medical Foundation New Scholar in Aging Award. Dr. Paternostro now holds adjunct faculty positions at Sanford Burnham Prebys Medical Discovery Institute and at the Department of Bioengineering, UC San Diego. His lab is located at Sanford Burnham Prebys.
Related Disease
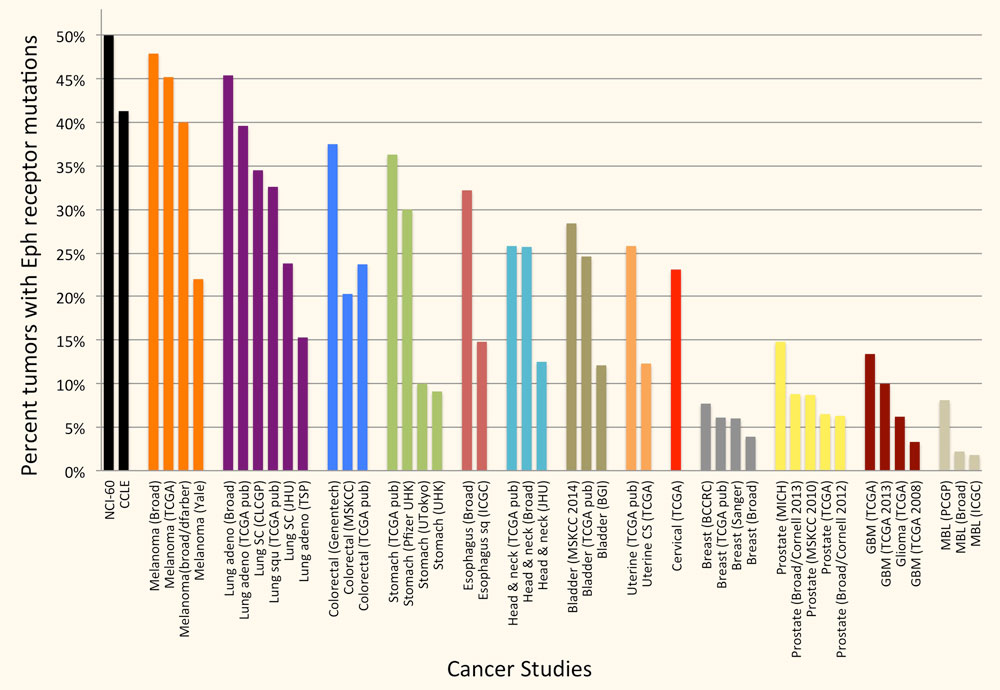
Leukemia/Lymphoma, Ovarian Cancer
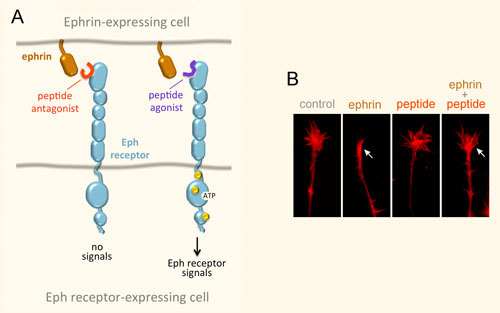
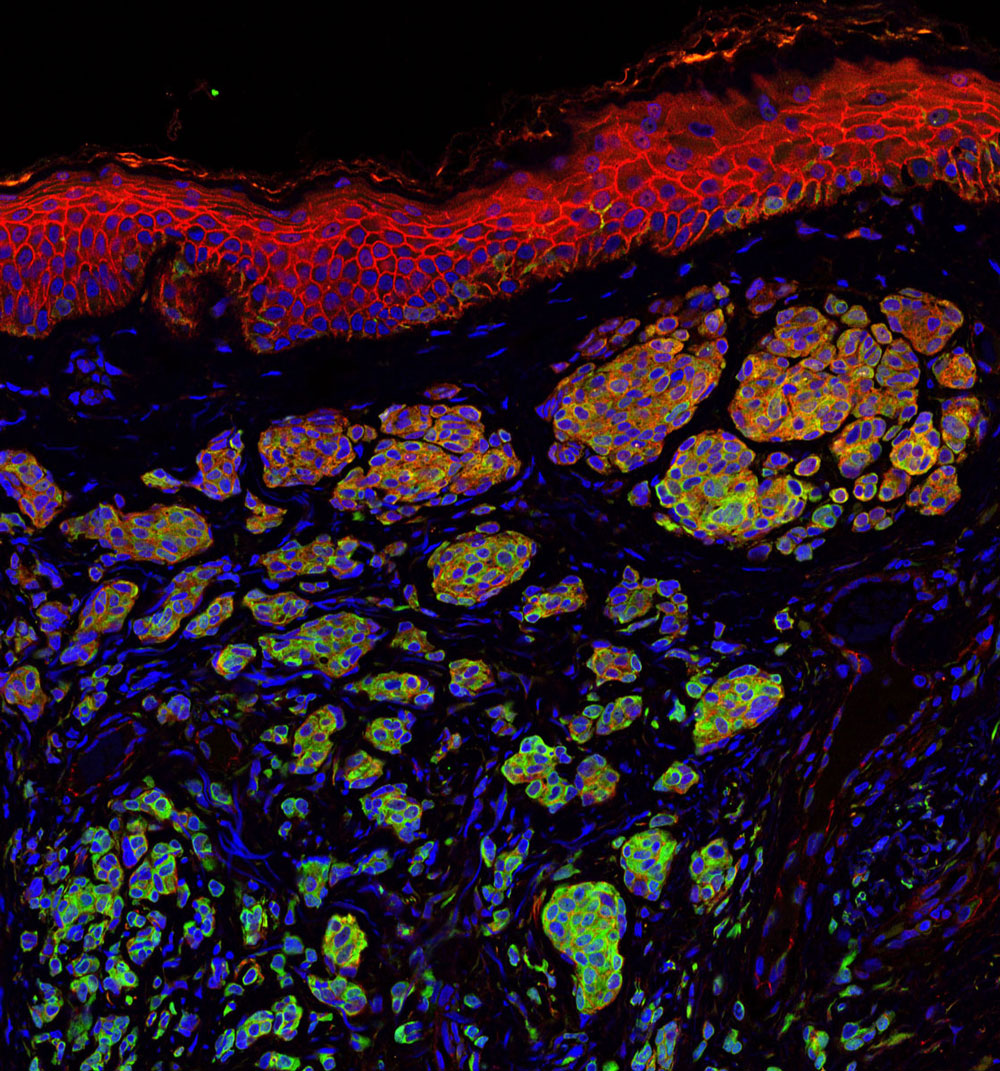
Our lab uses a systems biology approach to the study of complex diseases. Combined drug interventions are an increasingly common therapeutic approach to complex diseases, for example in cancer. Drugs are, however, usually developed individually and only later combined empirically in the clinic based on their known effects as single-therapy agents. We are interested in the problem of inducing selective cancer cell death. We have developed and validated search algorithms to discover optimal combinations of three or more drugs that would be infeasible to identify by fully combinatorial searches. In our procedure the optimization is not carried out in silico, but directly in an in vivo high-throughput system, where the response to therapeutic combinations is used as information to guide the system toward improved combinations using an iterative algorithm. System-wide molecular measurements (for example metabolomics and transcriptomics) and models can also be incorporated in these algorithms. It is useful to view the information processing by our experimental cellular systems as biological computations, since the algorithms we use are indeed often derived from algorithms that are implemented in silico in other scientific fields.
We also use the fruit fly (Drosophila) to study cardiac and metabolic alterations caused by aging and hypoxia, using high-throughput physiological measurements, NMR metabolomics and models of metabolism.
Our multi-disciplinary team is composed of biomedical and computational scientists, and we have close collaborations with physicists, engineers and bioengineers.